Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice
- PMID: 18835033
- PMCID: PMC2643030
- DOI: 10.1016/j.ccr.2008.09.001
Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice
Abstract
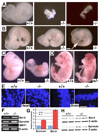
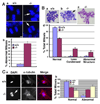
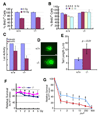
In lower eukaryotes, Sir2 serves as a histone deacetylase and is implicated in chromatin silencing, longevity, and genome stability. Here we mutated the Sirt1 gene, a homolog of yeast Sir2, in mice to study its function. We show that a majority of SIRT1 null embryos die between E9.5 and E14.5, displaying altered histone modification, impaired DNA damage response, and reduced ability to repair DNA damage. We demonstrate that Sirt1(+/-);p53(+/-) mice develop tumors in multiple tissues, whereas activation of SIRT1 by resveratrol treatment reduces tumorigenesis. Finally, we show that many human cancers exhibit reduced levels of SIRT1 compared to normal controls. Thus, SIRT1 may act as a tumor suppressor through its role in DNA damage response and genome integrity.
Figures




References
-
- Aggarwal BB, Bhardwaj A, Aggarwal RS, Seeram NP, Shishodia S, Takada Y. Role of resveratrol in prevention and therapy of cancer: preclinical and clinical studies. Anticancer Res. 2004;24:2783–2840. - PubMed
-
- Antoccia A, Ricordy R, Maraschio P, Prudente S, Tanzarella C. Chromosomal sensitivity to clastogenic agents and cell cycle perturbations in Nijmegen breakage syndrome lymphoblastoid cell lines. Int J Radiat Biol. 1997;71:41–49. - PubMed
-
- Aziz MH, Afaq F, Ahmad N. Prevention of ultraviolet-B radiation damage by resveratrol in mouse skin is mediated via modulation in survivin. Photochem Photobiol. 2005;81:25–31. - PubMed
-
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

