Interaction of the N- and C-terminal autoregulatory domains of FRL2 does not inhibit FRL2 activity
- PMID: 18835814
- PMCID: PMC2662283
- DOI: 10.1074/jbc.M803156200
Interaction of the N- and C-terminal autoregulatory domains of FRL2 does not inhibit FRL2 activity
Abstract
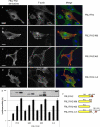
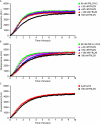
Formin homology proteins are a highly conserved family of cytoskeletal remodeling proteins best known for their ability to induce the formation of long unbranched actin filaments. They accomplish this by nucleating the de novo polymerization of F-actin and also by acting as F-actin barbed end "leaky cappers" that allow filament elongation while antagonizing the function of capping proteins. More recently, it has been reported that the FH2 domains of FRL1 and mDia2 and the plant formin AFH1 are able to bind and bundle actin filaments via distinct mechanisms. We find that like FRL1, FRL2 and FRL3 are also able to bind and bundle actin filaments. In the case of FRL3, this activity is dependent upon a proximal DAD/WH2-like domain that is found C-terminal to the FH2 domain. In addition, we show that, like other Diaphanous-related formins, FRL3 activity is subject to autoregulation mediated by the interaction between its N-terminal DID and C-terminal DAD. In contrast, the DID and DAD of FRL2 also interact in vivo and in vitro but without inhibiting FRL2 activity. These data suggest that current models describing DID/DAD autoregulation via steric hindrance of FH2 activity must be revised. Finally, unlike other formins, we find that the FH2 and N-terminal dimerization domains of FRL2 and FRL3 are able to form hetero-oligomers.
Figures






Similar articles
-
Assembly of filopodia by the formin FRL2 (FMNL3).Cytoskeleton (Hoboken). 2010 Dec;67(12):755-72. doi: 10.1002/cm.20485. Epub 2010 Nov 2. Cytoskeleton (Hoboken). 2010. PMID: 20862687 Free PMC article.
-
Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2.J Biol Chem. 2006 May 19;281(20):14383-92. doi: 10.1074/jbc.M510923200. Epub 2006 Mar 23. J Biol Chem. 2006. PMID: 16556604
-
Non diaphanous formin delphilin acts as a barbed end capping protein.Exp Cell Res. 2017 Aug 15;357(2):163-169. doi: 10.1016/j.yexcr.2017.05.014. Epub 2017 May 17. Exp Cell Res. 2017. PMID: 28527698
-
Review of the mechanism of processive actin filament elongation by formins.Cell Motil Cytoskeleton. 2009 Aug;66(8):606-17. doi: 10.1002/cm.20379. Cell Motil Cytoskeleton. 2009. PMID: 19459187 Free PMC article. Review.
-
Fifteen formins for an actin filament: a molecular view on the regulation of human formins.Biochim Biophys Acta. 2010 Feb;1803(2):152-63. doi: 10.1016/j.bbamcr.2010.01.014. Epub 2010 Jan 25. Biochim Biophys Acta. 2010. PMID: 20102729 Review.
Cited by
-
Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing.J Biol Chem. 2012 Oct 5;287(41):34234-45. doi: 10.1074/jbc.M112.365122. Epub 2012 Aug 9. J Biol Chem. 2012. PMID: 22879592 Free PMC article.
-
Critical roles for multiple formins during cardiac myofibril development and repair.Mol Biol Cell. 2014 Mar;25(6):811-27. doi: 10.1091/mbc.E13-08-0443. Epub 2014 Jan 15. Mol Biol Cell. 2014. PMID: 24430873 Free PMC article.
-
Identification of a short Spir interaction sequence at the C-terminal end of formin subgroup proteins.J Biol Chem. 2009 Sep 11;284(37):25324-33. doi: 10.1074/jbc.M109.030320. Epub 2009 Jul 15. J Biol Chem. 2009. PMID: 19605360 Free PMC article.
-
The formin DAD domain plays dual roles in autoinhibition and actin nucleation.Curr Biol. 2011 Mar 8;21(5):384-90. doi: 10.1016/j.cub.2011.01.047. Epub 2011 Feb 17. Curr Biol. 2011. PMID: 21333540 Free PMC article.
-
FMNL2 drives actin-based protrusion and migration downstream of Cdc42.Curr Biol. 2012 Jun 5;22(11):1005-12. doi: 10.1016/j.cub.2012.03.064. Epub 2012 May 17. Curr Biol. 2012. PMID: 22608513 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

