Chromosome 15q11-13 duplication syndrome brain reveals epigenetic alterations in gene expression not predicted from copy number
- PMID: 18835857
- PMCID: PMC2634820
- DOI: 10.1136/jmg.2008.061580
Chromosome 15q11-13 duplication syndrome brain reveals epigenetic alterations in gene expression not predicted from copy number
Abstract
Background: Chromosome 15q11-13 contains a cluster of imprinted genes essential for normal mammalian neurodevelopment. Deficiencies in paternal or maternal 15q11-13 alleles result in Prader-Willi or Angelman syndromes, respectively, and maternal duplications lead to a distinct condition that often includes autism. Overexpression of maternally expressed imprinted genes is predicted to cause 15q11-13-associated autism, but a link between gene dosage and expression has not been experimentally determined in brain.
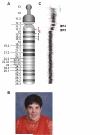
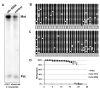
Methods: Postmortem brain tissue was obtained from a male with 15q11-13 hexasomy and a female with 15q11-13 tetrasomy. Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) was used to measure 10 15q11-13 transcripts in maternal 15q11-13 duplication, Prader-Willi syndrome, and control brain samples. Southern blot, bisulfite sequencing and fluorescence in situ hybridisation were used to investigate epigenetic mechanisms of gene regulation.
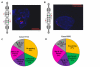
Results: Gene expression and DNA methylation correlated with parental gene dosage in the male 15q11-13 duplication sample with severe cognitive impairment and seizures. Strikingly, the female with autism and milder Prader-Willi-like characteristics demonstrated unexpected deficiencies in the paternally expressed transcripts SNRPN, NDN, HBII85, and HBII52 and unchanged levels of maternally expressed UBE3A compared to controls. Paternal expression abnormalities in the female duplication sample were consistent with elevated DNA methylation of the 15q11-13 imprinting control region (ICR). Expression of non-imprinted 15q11-13 GABA receptor subunit genes was significantly reduced specifically in the female 15q11-13 duplication brain without detectable GABRB3 methylation differences.
Conclusion: Our findings suggest that genetic copy number changes combined with additional genetic or environmental influences on epigenetic mechanisms impact outcome and clinical heterogeneity of 15q11-13 duplication syndromes.
Figures


References
-
- Schroer RJ, Phelan MC, Michaelis RC, et al. Autism and maternally derived aberrations of chromosome 15q. Am J Med Genet. 1998;76(4):327–36. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
