A genetic timer through noise-induced stabilization of an unstable state
- PMID: 18836072
- PMCID: PMC2560995
- DOI: 10.1073/pnas.0806349105
A genetic timer through noise-induced stabilization of an unstable state
Abstract
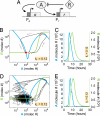
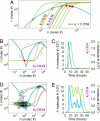
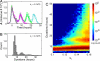
Stochastic fluctuations affect the dynamics of biological systems. Typically, such noise causes perturbations that can permit genetic circuits to escape stable states, triggering, for example, phenotypic switching. In contrast, studies have shown that noise can surprisingly also generate new states, which exist solely in the presence of fluctuations. In those instances noise is supplied externally to the dynamical system. Here, we present a mechanism in which noise intrinsic to a simple genetic circuit effectively stabilizes a deterministically unstable state. Furthermore, this noise-induced stabilization represents a unique mechanism for a genetic timer. Specifically, we analyzed the effect of noise intrinsic to a prototypical two-component gene-circuit architecture composed of interacting positive and negative feedback loops. Genetic circuits with this topology are common in biology and typically regulate cell cycles and circadian clocks. These systems can undergo a variety of bifurcations in response to parameter changes. Simulations show that near one such bifurcation, noise induces oscillations around an unstable spiral point and thus effectively stabilizes this unstable fixed point. Because of the periodicity of these oscillations, the lifetime of the noise-dependent stabilization exhibits a polymodal distribution with multiple, well defined, and regularly spaced peaks. Therefore, the noise-induced stabilization presented here constitutes a minimal mechanism for a genetic circuit to function as a timer that could be used in the engineering of synthetic circuits.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
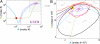
References
-
- Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38(6):636–643. - PubMed
-
- Blake WJ, et al. Noise in eukaryotic gene expression. Nature. 2003;422(6932):633–637. - PubMed
-
- Elowitz MB, et al. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. - PubMed
-
- Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–846. - PubMed
-
- Ozbudak EM, et al. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31(1):69–73. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

