Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5
- PMID: 18836439
- PMCID: PMC2635539
- DOI: 10.1038/ncb1786
Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5
Abstract
Mouse ES cells can differentiate into all three germ layers of the embryo but are generally excluded from the trophoblast lineage. Here we show that ES cells deficient in DNA methylation can differentiate efficiently into trophoblast derivatives. In a genome-wide screen we identified the transcription factor Elf5 as methylated and repressed in ES cells, and hypomethylated and expressed in TS and methylation-deficient ES cells. Elf5 creates a positive-feedback loop with the TS cell determinants Cdx2 and Eomes that is restricted to the trophoblast lineage by epigenetic regulation of Elf5. Importantly, the late-acting function of Elf5 allows initial plasticity and regulation in the early blastocyst. Thus, Elf5 functions as a gatekeeper, downstream of initial lineage determination, to reinforce commitment to the trophoblast lineage or to abort this pathway in epiblast cells. This epigenetic restriction of cell lineage fate provides a molecular mechanism for Waddington's concept of canalization of developmental pathways.
Figures

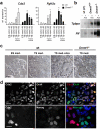
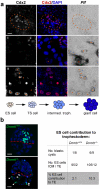

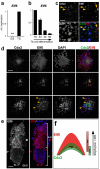
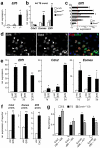

References
-
- Gardner RL. Clonal analysis of early mammalian development. Philos Trans R Soc Lond B Biol Sci. 1985;312:163–78. - PubMed
-
- Dyce J, George M, Goodall H, Fleming TP. Do trophectoderm and inner cell mass cells in the mouse blastocyst maintain discrete lineages? Development. 1987;100:685–98. - PubMed
-
- Fleming TP. A quantitative analysis of cell allocation to trophectoderm and inner cell mass in the mouse blastocyst. Dev Biol. 1987;119:520–31. - PubMed
-
- Johnson MH, McConnell JM. Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev Biol. 2004;15:583–97. - PubMed
-
- Bradley A, Robertson E. Embryo-derived stem cells: a tool for elucidating the developmental genetics of the mouse. Curr Top Dev Biol. 1986;20:357–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0700098/MRC_/Medical Research Council/United Kingdom
- G0700098(81308)/MRC_/Medical Research Council/United Kingdom
- BBS/E/B/0000M100/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0400154/MRC_/Medical Research Council/United Kingdom
- BBS/E/B/0000C225/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/B/0000L709/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G120/824(65167)/MRC_/Medical Research Council/United Kingdom
- G0300723/MRC_/Medical Research Council/United Kingdom
- G120/824/MRC_/Medical Research Council/United Kingdom
- G0800784/MRC_/Medical Research Council/United Kingdom
- G0400154(69909)/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

