Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip
- PMID: 18836445
- PMCID: PMC2691688
- DOI: 10.1038/ng.242
Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip
Abstract
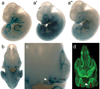
Previously we have shown that nonsyndromic cleft lip with or without cleft palate (NSCL/P) is strongly associated with SNPs in IRF6 (interferon regulatory factor 6). Here, we use multispecies sequence comparisons to identify a common SNP (rs642961, G>A) in a newly identified IRF6 enhancer. The A allele is significantly overtransmitted (P = 1 x 10(-11)) in families with NSCL/P, in particular those with cleft lip but not cleft palate. Further, there is a dosage effect of the A allele, with a relative risk for cleft lip of 1.68 for the AG genotype and 2.40 for the AA genotype. EMSA and ChIP assays demonstrate that the risk allele disrupts the binding site of transcription factor AP-2alpha and expression analysis in the mouse localizes the enhancer activity to craniofacial and limb structures. Our findings place IRF6 and AP-2alpha in the same developmental pathway and identify a high-frequency variant in a regulatory element contributing substantially to a common, complex disorder.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG003988/HG/NHGRI NIH HHS/United States
- R01 ES010876/ES/NIEHS NIH HHS/United States
- R37 DE008559/DE/NIDCR NIH HHS/United States
- P30 ES005605/ES/NIEHS NIH HHS/United States
- R01-HG003988/HG/NHGRI NIH HHS/United States
- P50 DE009170/DE/NIDCR NIH HHS/United States
- R01 DE013513/DE/NIDCR NIH HHS/United States
- R37 DE08559/DE/NIDCR NIH HHS/United States
- 1 UL1 RR024979-01/RR/NCRR NIH HHS/United States
- R01-CA73612/CA/NCI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- P30 ES05605/ES/NIEHS NIH HHS/United States
- P50 DE016215/DE/NIDCR NIH HHS/United States
- R01 CA073612/CA/NCI NIH HHS/United States
- MC_U127561093/MRC_/Medical Research Council/United Kingdom
- R01 DE016148/DE/NIDCR NIH HHS/United States
- R01-DE13513/DE/NIDCR NIH HHS/United States
- R01 DE008559/DE/NIDCR NIH HHS/United States
- T32 CA078586/CA/NCI NIH HHS/United States
- P50 DE16215/DE/NIDCR NIH HHS/United States
- UL1 RR024979/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

