Regulation of a remote Shh forebrain enhancer by the Six3 homeoprotein
- PMID: 18836447
- PMCID: PMC2648611
- DOI: 10.1038/ng.230
Regulation of a remote Shh forebrain enhancer by the Six3 homeoprotein
Abstract
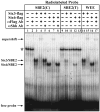
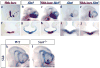
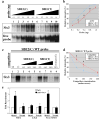
In humans, SHH haploinsufficiency results in holoprosencephaly (HPE), a defect in anterior midline formation. Despite the importance of maintaining SHH transcript levels above a critical threshold, we know little about the upstream regulators of SHH expression in the forebrain. Here we describe a rare nucleotide variant located 460 kb upstream of SHH in an individual with HPE that resulted in the loss of Shh brain enhancer-2 (SBE2) activity in the hypothalamus of transgenic mouse embryos. Using a DNA affinity-capture assay, we screened the SBE2 sequence for DNA-binding proteins and identified members of the Six3 and Six6 homeodomain family as candidate regulators of Shh transcription. Six3 showed reduced binding affinity for the mutant compared to the wild-type SBE2 sequence. Moreover, Six3 with HPE-causing alterations failed to bind and activate SBE2. These data suggest a direct link between Six3 and Shh regulation during normal forebrain development and in the pathogenesis of HPE.
Figures


References
-
- Chiang C, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. - PubMed
-
- Britto J, Tannahill D, Keynes R. A critical role for sonic hedgehog signaling in the early expansion of the developing brain. Nat Neurosci. 2002;5:103–10. - PubMed
-
- Marcucio RS, Cordero DR, Hu D, Helms JA. Molecular interactions coordinating the development of the forebrain and face. Dev Biol. 2005;284:48–61. - PubMed
-
- Roessler E, et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

