The Caenorhabditis chemoreceptor gene families
- PMID: 18837995
- PMCID: PMC2576165
- DOI: 10.1186/1741-7007-6-42
The Caenorhabditis chemoreceptor gene families
Abstract
Background: Chemoreceptor proteins mediate the first step in the transduction of environmental chemical stimuli, defining the breadth of detection and conferring stimulus specificity. Animal genomes contain families of genes encoding chemoreceptors that mediate taste, olfaction, and pheromone responses. The size and diversity of these families reflect the biology of chemoperception in specific species.
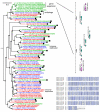
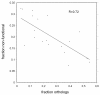
Results: Based on manual curation and sequence comparisons among putative G-protein-coupled chemoreceptor genes in the nematode Caenorhabditis elegans, we identified approximately 1300 genes and 400 pseudogenes in the 19 largest gene families, most of which fall into larger superfamilies. In the related species C. briggsae and C. remanei, we identified most or all genes in each of the 19 families. For most families, C. elegans has the largest number of genes and C. briggsae the smallest number, suggesting changes in the importance of chemoperception among the species. Protein trees reveal family-specific and species-specific patterns of gene duplication and gene loss. The frequency of strict orthologs varies among the families, from just over 50% in two families to less than 5% in three families. Several families include large species-specific expansions, mostly in C. elegans and C. remanei.
Conclusion: Chemoreceptor gene families in Caenorhabditis species are large and evolutionarily dynamic as a result of gene duplication and gene loss. These dynamics shape the chemoreceptor gene complements in Caenorhabditis species and define the receptor space available for chemosensory responses. To explain these patterns, we propose the gray pawn hypothesis: individual genes are of little significance, but the aggregate of a large number of diverse genes is required to cover a large phenotype space.
Figures
Similar articles
-
Adaptive evolution in the SRZ chemoreceptor families of Caenorhabditis elegans and Caenorhabditis briggsae.Proc Natl Acad Sci U S A. 2005 Mar 22;102(12):4476-81. doi: 10.1073/pnas.0406469102. Epub 2005 Mar 10. Proc Natl Acad Sci U S A. 2005. PMID: 15761060 Free PMC article.
-
Two large families of chemoreceptor genes in the nematodes Caenorhabditis elegans and Caenorhabditis briggsae reveal extensive gene duplication, diversification, movement, and intron loss.Genome Res. 1998 May;8(5):449-63. doi: 10.1101/gr.8.5.449. Genome Res. 1998. PMID: 9582190
-
The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses.Genome Res. 2000 Feb;10(2):192-203. doi: 10.1101/gr.10.2.192. Genome Res. 2000. PMID: 10673277
-
The putative chemoreceptor families of C. elegans.WormBook. 2006 Jan 6:1-12. doi: 10.1895/wormbook.1.66.1. WormBook. 2006. PMID: 18050473 Free PMC article. Review.
-
Chemosensory signaling in C. elegans.Bioessays. 1999 Dec;21(12):1011-20. doi: 10.1002/(SICI)1521-1878(199912)22:1<1011::AID-BIES5>3.0.CO;2-V. Bioessays. 1999. PMID: 10580986 Review.
Cited by
-
Neuropeptide GPCRs in C. elegans.Front Endocrinol (Lausanne). 2012 Dec 21;3:167. doi: 10.3389/fendo.2012.00167. eCollection 2012. Front Endocrinol (Lausanne). 2012. PMID: 23267347 Free PMC article.
-
Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus.PLoS Pathog. 2011 Sep;7(9):e1002219. doi: 10.1371/journal.ppat.1002219. Epub 2011 Sep 1. PLoS Pathog. 2011. PMID: 21909270 Free PMC article.
-
Multigenic natural variation underlies Caenorhabditis elegans olfactory preference for the bacterial pathogen Serratia marcescens.G3 (Bethesda). 2014 Feb 19;4(2):265-76. doi: 10.1534/g3.113.008649. G3 (Bethesda). 2014. PMID: 24347628 Free PMC article.
-
Gene-environment and protein-degradation signatures characterize genomic and phenotypic diversity in wild Caenorhabditis elegans populations.BMC Biol. 2013 Aug 19;11:93. doi: 10.1186/1741-7007-11-93. BMC Biol. 2013. PMID: 23957880 Free PMC article.
-
Genetic and functional diversification of chemosensory pathway receptors in mosquito-borne filarial nematodes.PLoS Biol. 2020 Jun 8;18(6):e3000723. doi: 10.1371/journal.pbio.3000723. eCollection 2020 Jun. PLoS Biol. 2020. PMID: 32511224 Free PMC article.
References
-
- Bargmann CI. Chemosensation in C. elegans. Wormbook. 2006 http://www.wormbook.org/chapters/www_chemosensation/chemosensation.html - PMC - PubMed
-
- Robertson HM. Two large families of chemoreceptor genes in the nematodes Caenorhabditis elegans and Caenorhabditis briggsae reveal extensive gene duplication, diversification, movement, and intron loss. Genome Res. 1998;8:449–463. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

