Notch activates cell cycle reentry and progression in quiescent cardiomyocytes
- PMID: 18838555
- PMCID: PMC2557048
- DOI: 10.1083/jcb.200806104
Notch activates cell cycle reentry and progression in quiescent cardiomyocytes
Abstract
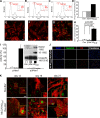
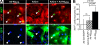
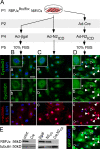
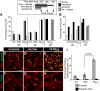
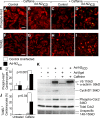
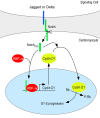
The inability of heart muscle to regenerate by replication of existing cardiomyocytes has engendered considerable interest in identifying developmental or other stimuli capable of sustaining the proliferative capacity of immature cardiomyocytes or stimulating division of postmitotic cardiomyocytes. Here, we demonstrate that reactivation of Notch signaling causes embryonic stem cell-derived and neonatal ventricular cardiomyocytes to enter the cell cycle. The proliferative response of neonatal ventricular cardiomyocytes declines as they mature, such that late activation of Notch triggers the DNA damage checkpoint and G2/M interphase arrest. Notch induces recombination signal-binding protein 1 for Jkappa (RBP-Jkappa)-dependent expression of cyclin D1 but, unlike other inducers, also shifts its subcellular distribution from the cytosol to the nucleus. Nuclear localization of cyclin D1 is independent of RBP-Jkappa. Thus, the influence of Notch on nucleocytoplasmic localization of cyclin D1 is an unanticipated property of the Notch intracellular domain that is likely to regulate the cell cycle in multiple contexts, including tumorigenesis as well as cardiogenesis.
Figures



References
-
- Ahuja, P., E. Perriard, J.C. Perriard, and E. Ehler. 2004. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J. Cell Sci. 117:3295–3306. - PubMed
-
- Barcova, M., V.M. Campa, and M. Mercola. 2007. Human embryonic stem cell cardiogenesis. In Human Stem Cell Manual: a Laboratory Guide. J.F. Loring, R.L. Wesselschmidt, and P.H. Schwartz, editors. Elsevier/Academic Press, Amsterdam. 227–237.
-
- Belka, C. 2006. The fate of irradiated tumor cells. Oncogene. 25:969–971. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

