Selected replicon variants with low-level in vitro resistance to the hepatitis C virus NS5B polymerase inhibitor PSI-6130 lack cross-resistance with R1479
- PMID: 18838588
- PMCID: PMC2592884
- DOI: 10.1128/AAC.00444-08
Selected replicon variants with low-level in vitro resistance to the hepatitis C virus NS5B polymerase inhibitor PSI-6130 lack cross-resistance with R1479
Abstract
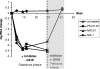
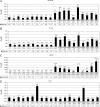
PSI-6130 (beta-D-2'-deoxy-2'-fluoro-2'-C-methylcytidine) is a selective inhibitor of hepatitis C virus (HCV) replication that targets the NS5B polymerase. R7128, the prodrug of PSI-6130, has shown antiviral efficacy in patients chronically infected with HCV genotype 1a (GT-1a) and GT-1b. We observed that the compound exhibited potent in vitro activity against laboratory-optimized HCV replicons as well as against a panel of replicons containing NS5B HCV polymerases derived from GT-1a and GT-1b clinical isolates. We used the HCV replicon cell system to examine the emergence of variants with reduced sensitivity to PSI-6130. Short-term treatment of cells harboring the HCV subgenomic replicon with PSI-6130 cleared the replicon without generating resistant variants. Long-term culture of the cells under the compound selection generated the S282T substitution in a complex pattern with other amino acid substitutions in the NS5B polymerase. The presence of the coselected substitutions did not increase the moderate three- to sixfold loss of sensitivity to PSI-6130 mediated by the S282T substitution; however, their presence enhanced the replication capacity compared to the replication levels seen with the S282T substitution alone. We also observed a lack of cross-resistance between PSI-6130 and R1479 and demonstrated that long-term culture selection with PSI-6130 in replicon cells harboring preexisting mutations resistant to R1479 (S96T/N142T) results in the emergence of the S282T substitution and the reversion of S96T to wild-type serine. In conclusion, PSI-6130 presents a high barrier to resistance selection in vitro, selects for variants exhibiting only low-level resistance, and lacks cross-resistance with R1479, supporting the continued development of the prodrug R7128 as a therapeutic agent for the treatment of HCV infection.
Figures





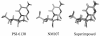
References
-
- Afdhal, N., M. Rodriguez-Torres, E. Lawitz, E. Godofsky, G. Chao, B. Fielman, S. Knox, and N. Brown. 2005. Enhanced antiviral efficacy for valopicitabine (NM283) plus Peg-interferon in hepatitis C patients with HCV genotype-1 infection: results of a phase IIa multicenter trial, abstr. LB03. Abstr. 40th Annu. Meet. Eur. Assoc. Study Liver, Paris, France, 13 to 17 April, 2005.
-
- Beaulieu, P. L. 2006. Finger loop inhibitors of the HCV NS5B polymerase: discovery and prospects for new HCV therapy. Curr. Opin. Drug Discov. Devel. 9:618-626. - PubMed
-
- Beaulieu, P. L. 2007. Non-nucleoside inhibitors of the HCV NS5B polymerase: progress in the discovery and development of novel agents for the treatment of HCV infections. Curr. Opin. Investig. Drugs 8:614-634. - PubMed
-
- Beaulieu, P. L., M. Bos, Y. Bousquet, P. DeRoy, G. Fazal, J. Gauthier, J. Gillard, S. Goulet, G. McKercher, M. A. Poupart, S. Valois, and G. Kukolj. 2004. Non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase: discovery of benzimidazole 5-carboxylic amide derivatives with low-nanomolar potency. Bioorg. Med. Chem. Lett. 14:967-971. - PubMed
-
- Beaulieu, P. L., J. Gillard, D. Bykowski, C. Brochu, N. Dansereau, J. S. Duceppe, B. Hache, A. Jakalian, L. Lagace, S. LaPlante, G. McKercher, E. Moreau, S. Perreault, T. Stammers, L. Thauvette, J. Warrington, and G. Kukolj. 2006. Improved replicon cellular activity of non-nucleoside allosteric inhibitors of HCV NS5B polymerase: from benzimidazole to indole scaffolds. Bioorg. Med. Chem. Lett. 16:4987-4993. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

