Human alkyladenine DNA glycosylase employs a processive search for DNA damage
- PMID: 18839966
- PMCID: PMC2702167
- DOI: 10.1021/bi801046y
Human alkyladenine DNA glycosylase employs a processive search for DNA damage
Abstract
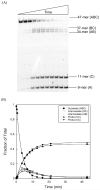
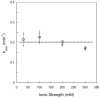
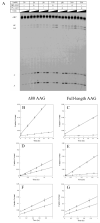
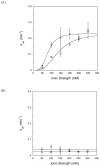
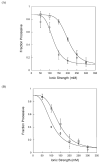
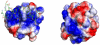
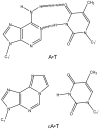
DNA repair proteins conduct a genome-wide search to detect and repair sites of DNA damage wherever they occur. Human alkyladenine DNA glycosylase (AAG) is responsible for recognizing a variety of base lesions, including alkylated and deaminated purines, and initiating their repair via the base excision repair pathway. We have investigated the mechanism by which AAG locates sites of damage using an oligonucleotide substrate containing two sites of DNA damage. This substrate was designed so that AAG randomly binds to either of the two lesions. AAG-catalyzed base excision creates a repair intermediate, and the subsequent partitioning between dissociation and diffusion to the second site can be quantified from the rates of formation of the different products. Our results demonstrate that AAG has the ability to slide for short distances along DNA at physiological salt concentrations. The processivity of AAG decreases with increasing ionic strength to become fully distributive at high ionic strengths, suggesting that electrostatic interactions between the negatively charged DNA and the positively charged DNA binding surface are important for nonspecific DNA binding. Although the amino terminus of the protein is dispensable for glycosylase activity at a single site, we find that deletion of the 80 amino-terminal amino acids significantly decreases the processivity of AAG. These observations support the idea that diffusion on undamaged DNA contributes to the search for sites of DNA damage.
Figures
References
-
- Ye N, Holmquist GP, O’Connor TR. Heterogeneous repair of N-methylpurines at the nucleotide level in normal human cells. J Mol Biol. 1998;284:269–85. - PubMed
-
- Cappelli E, Hazra T, Hill JW, Slupphaug G, Bogliolo M, Frosina G. Rates of base excision repair are not solely dependent on levels of initiating enzymes. Carcinogenesis. 2001;22:387–93. - PubMed
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. - PubMed
-
- O’Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem. 2004;279:9750–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

