High-resolution array CGH clarifies events occurring on 8p in carcinogenesis
- PMID: 18840272
- PMCID: PMC2576333
- DOI: 10.1186/1471-2407-8-288
High-resolution array CGH clarifies events occurring on 8p in carcinogenesis
Abstract
Background: Rearrangement of the short arm of chromosome 8 (8p) is very common in epithelial cancers such as breast cancer. Usually there is an unbalanced translocation breakpoint in 8p12 (29.7 Mb - 38.5 Mb) with loss of distal 8p, sometimes with proximal amplification of 8p11-12. Rearrangements in 8p11-12 have been investigated using high-resolution array CGH, but the first 30 Mb of 8p are less well characterised, although this region contains several proposed tumour suppressor genes.
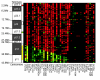
Methods: We analysed the whole of 8p by array CGH at tiling-path BAC resolution in 32 breast and six pancreatic cancer cell lines. Regions of recurrent rearrangement distal to 8p12 were further characterised, using regional fosmid arrays. FISH, and quantitative RT-PCR on over 60 breast tumours validated the existence of similar events in primary material.
Results: We confirmed that 8p is usually lost up to at least 30 Mb, but a few lines showed focal loss or copy number steps within this region. Three regions showed rearrangements common to at least two cases: two regions of recurrent loss and one region of amplification. Loss within 8p23.3 (0 Mb - 2.2 Mb) was found in six cell lines. Of the genes always affected, ARHGEF10 showed a point mutation of the remaining normal copies in the DU4475 cell line. Deletions within 12.7 Mb - 19.1 Mb in 8p22, in two cases, affected TUSC3. A novel amplicon was found within 8p21.3 (19.1 Mb - 23.4 Mb) in two lines and one of 98 tumours.
Conclusion: The pattern of rearrangements seen on 8p may be a consequence of the high density of potential targets on this chromosome arm, and ARHGEF10 may be a new candidate tumour suppressor gene.
Figures




References
-
- Loo LW, Grove DI, Williams EM, Neal CL, Cousens LA, Schubert EL, Holcomb IN, Massa HF, Glogovac J, Li CI, Malone KE, Daling JR, Delrow JJ, Trask BJ, Hsu L, Porter PL. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. - DOI - PubMed
-
- Bashyam MD, Bair R, Kim YH, Wang P, Hernandez-Boussard T, Karikari CA, Tibshirani R, Maitra A, Pollack JR. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia. 2005;7:556–562. doi: 10.1593/neo.04586. - DOI - PMC - PubMed
-
- Emi M, Fujiwara Y, Nakajima T, Tsuchiya E, Tsuda H, Hirohashi S, Maeda Y, Tsuruta K, Miyaki M, Nakamura Y. Frequent loss of heterozygosity for loci on chromosome 8p in hepatocellular carcinoma, colorectal cancer, and lung cancer. Cancer Res. 1992;52:5368–5372. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

