Exploring RNA transcription and turnover in vivo by using click chemistry
- PMID: 18840688
- PMCID: PMC2572917
- DOI: 10.1073/pnas.0808480105
Exploring RNA transcription and turnover in vivo by using click chemistry
Abstract
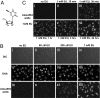
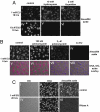
We describe a chemical method to detect RNA synthesis in cells, based on the biosynthetic incorporation of the uridine analog 5-ethynyluridine (EU) into newly transcribed RNA, on average once every 35 uridine residues in total RNA. EU-labeled cellular RNA is detected quickly and with high sensitivity by using a copper (I)-catalyzed cycloaddition reaction (often referred to as "click" chemistry) with fluorescent azides, followed by microscopic imaging. We demonstrate the use of this method in cultured cells, in which we examine the turnover of bulk RNA after EU pulses of varying lengths. We also use EU to assay transcription rates of various tissues in whole animals, both on sections and by whole-mount staining. We find that total transcription rates vary greatly among different tissues and among different cell types within organs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Haukenes G, et al. Labeling of RNA transcripts of eukaryotic cells in culture with BrUTP using a liposome transfection reagent (DOTAP) BioTechniques. 1997;22:308–312. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

