Global reorganization of replication domains during embryonic stem cell differentiation
- PMID: 18842067
- PMCID: PMC2561079
- DOI: 10.1371/journal.pbio.0060245
Global reorganization of replication domains during embryonic stem cell differentiation
Abstract
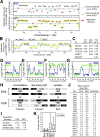
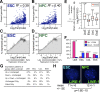
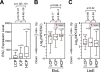
DNA replication in mammals is regulated via the coordinate firing of clusters of replicons that duplicate megabase-sized chromosome segments at specific times during S-phase. Cytogenetic studies show that these "replicon clusters" coalesce as subchromosomal units that persist through multiple cell generations, but the molecular boundaries of such units have remained elusive. Moreover, the extent to which changes in replication timing occur during differentiation and their relationship to transcription changes has not been rigorously investigated. We have constructed high-resolution replication-timing profiles in mouse embryonic stem cells (mESCs) before and after differentiation to neural precursor cells. We demonstrate that chromosomes can be segmented into multimegabase domains of coordinate replication, which we call "replication domains," separated by transition regions whose replication kinetics are consistent with large originless segments. The molecular boundaries of replication domains are remarkably well conserved between distantly related ESC lines and induced pluripotent stem cells. Unexpectedly, ESC differentiation was accompanied by the consolidation of smaller differentially replicating domains into larger coordinately replicated units whose replication time was more aligned to isochore GC content and the density of LINE-1 transposable elements, but not gene density. Replication-timing changes were coordinated with transcription changes for weak promoters more than strong promoters, and were accompanied by rearrangements in subnuclear position. We conclude that replication profiles are cell-type specific, and changes in these profiles reveal chromosome segments that undergo large changes in organization during differentiation. Moreover, smaller replication domains and a higher density of timing transition regions that interrupt isochore replication timing define a novel characteristic of the pluripotent state.
Conflict of interest statement
Figures





Comment in
-
Tying replication to cell identity.Nat Rev Mol Cell Biol. 2013 Jun;14(6):326. doi: 10.1038/nrm3593. Nat Rev Mol Cell Biol. 2013. PMID: 23698578 No abstract available.
References
-
- Gilbert DM, Gasser SM. Nuclear structure and DNA replication. In: DePamphilis ML, editor. DNA replication and human disease. Cold Spring Harbor (New York): Cold Spring Harbor Press; 2006. pp. 175–196.
-
- Sadoni N, Cardoso MC, Stelzer EH, Leonhardt H, Zink D. Stable chromosomal units determine the spatial and temporal organization of DNA replication. J Cell Sci. 2004;117:5353–5365. - PubMed
-
- Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1-phase. Mol Cell. 1999;4:983–993. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

