Plant ion channels: gene families, physiology, and functional genomics analyses
- PMID: 18842100
- PMCID: PMC4790454
- DOI: 10.1146/annurev.physiol.010908.163204
Plant ion channels: gene families, physiology, and functional genomics analyses
Abstract
Distinct potassium, anion, and calcium channels in the plasma membrane and vacuolar membrane of plant cells have been identified and characterized by patch clamping. Primarily owing to advances in Arabidopsis genetics and genomics, and yeast functional complementation, many of the corresponding genes have been identified. Recent advances in our understanding of ion channel genes that mediate signal transduction and ion transport are discussed here. Some plant ion channels, for example, ALMT and SLAC anion channel subunits, are unique. The majority of plant ion channel families exhibit homology to animal genes; such families include both hyperpolarization- and depolarization-activated Shaker-type potassium channels, CLC chloride transporters/channels, cyclic nucleotide-gated channels, and ionotropic glutamate receptor homologs. These plant ion channels offer unique opportunities to analyze the structural mechanisms and functions of ion channels. Here we review gene families of selected plant ion channel classes and discuss unique structure-function aspects and their physiological roles in plant cell signaling and transport.
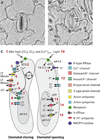
Figures

References
-
- Hope AB, Findlay GP. The action potential in Chara. Plant Cell Physiol. 1962;5:377–379.
-
- Hodick D, Sievers A. The action potential of Dionaea muscipula Ellis. Planta. 1988;174:8–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

