Preferred and avoided codon pairs in three domains of life
- PMID: 18842120
- PMCID: PMC2585594
- DOI: 10.1186/1471-2164-9-463
Preferred and avoided codon pairs in three domains of life
Abstract
Background: Alternative synonymous codons are not used with equal frequencies. In addition, the contexts of codons - neighboring nucleotides and neighboring codons - can have certain patterns. The codon context can influence both translational accuracy and elongation rates. However, it is not known how strong or conserved the codon context preferences in different organisms are. We analyzed 138 organisms (bacteria, archaea and eukaryotes) to find conserved patterns of codon pairs.
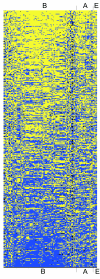
Results: After removing the effects of single codon usage and dipeptide biases we discovered a set of neighboring codons for which avoidances or preferences were conserved in all three domains of life. Such biased codon pairs could be divided into subtypes on the basis of the nucleotide patterns that influence the bias. The most frequently avoided type of codon pair was nnUAnn. We discovered that 95.7% of avoided nnUAnn type patterns contain out-frame UAA or UAG triplets on the sense and/or antisense strand. On average, nnUAnn codon pairs are more frequently avoided in ORFeomes than in genomes. Thus we assume that translational selection plays a major role in the avoidance of these codon pairs. Among the preferred codon pairs, nnGCnn was the major type.
Conclusion: Translational selection shapes codon pair usage in protein coding sequences by rules that are common to all three domains of life. The most frequently avoided codon pairs contain the patterns nnUAnn, nnGGnn, nnGnnC, nnCGCn, GUCCnn, CUCCnn, nnCnnA or UUCGnn. The most frequently preferred codon pairs contain the patterns nnGCnn, nnCAnn or nnUnCn.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

