Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner
- PMID: 18842586
- PMCID: PMC3259897
- DOI: 10.1074/jbc.M803219200
Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner
Abstract
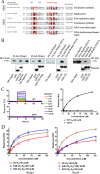
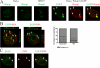
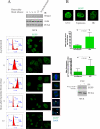
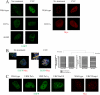
Wrnip1 (Werner helicase-interacting protein 1) has been implicated in the bypass of stalled replication forks in bakers' yeast. However, the function(s) of human Wrnip1 has remained elusive so far. Here we report that Wrnip1 is distributed inside heterogeneous structures detectable in nondamaged cells throughout the cell cycle. In an attempt to characterize these structures, we found that Wrnip1 resides in DNA replication factories. Upon treatments that stall replication forks, such as UVC light, the amount of chromatin-bound Wrnip1 and the number of foci significantly increase, further implicating Wrnip1 in DNA replication. Interestingly, the nuclear pattern of Wrnip1 appears to extend to a broader landscape, as it can be detected in promyelocytic leukemia nuclear bodies. The presence of Wrnip1 into these heterogeneous subnuclear structures requires its ubiquitin-binding zinc finger (UBZ) domain, which is able to interact with different ubiquitin (Ub) signals, including mono-Ub and chains linked via lysine 48 and 63. Moreover, the oligomerization of Wrnip1 mediated by its C terminus is also important for proper subnuclear localization. Our study is the first to reveal the composite and regulated topography of Wrnip1 in the human nucleus, highlighting its potential role in replication and other nuclear transactions.
Figures

References
-
- Branzei, D., and Foiani, M. (2008) Nat. Rev. Mol. Cell Biol. 9297 -308 - PubMed
-
- Moldovan, G. L., Pfander, B., and Jentsch, S. (2007) Cell 129665 -679 - PubMed
-
- Hickson, I. D. (2003) Nat. Rev. Cancer 3169 -178 - PubMed
-
- Kudlow, B. A., Kennedy, B. K., and Monnat, R. J., Jr. (2007) Nat. Rev. Mol. Cell Biol. 8 394-404 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

