Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans
- PMID: 18842709
- PMCID: PMC2593359
- DOI: 10.1128/JVI.01563-08
Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans
Abstract
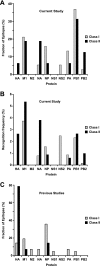
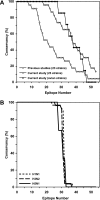
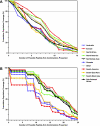
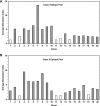
Continuing antigenic drift allows influenza viruses to escape antibody-mediated recognition, and as a consequence, the vaccine currently in use needs to be altered annually. Highly conserved epitopes recognized by effector T cells may represent an alternative approach for the generation of a more universal influenza virus vaccine. Relatively few highly conserved epitopes are currently known in humans, and relatively few epitopes have been identified from proteins other than hemagglutinin and nucleoprotein. This prompted us to perform a study aimed at identifying a set of human T-cell epitopes that would provide broad coverage against different virus strains and subtypes. To provide coverage across different ethnicities, seven different HLA supertypes were considered. More than 4,000 peptides were selected from a panel of 23 influenza A virus strains based on predicted high-affinity binding to HLA class I or class II and high conservancy levels. Peripheral blood mononuclear cells from 44 healthy human blood donors were tested for reactivity against HLA-matched peptides by using gamma interferon enzyme-linked immunospot assays. Interestingly, we found that PB1 was the major target for both CD4(+) and CD8(+) T-cell responses. The 54 nonredundant epitopes (38 class I and 16 class II) identified herein provided high coverage among different ethnicities, were conserved in the majority of the strains analyzed, and were consistently recognized in multiple individuals. These results enable further functional studies of T-cell responses during influenza virus infection and provide a potential base for the development of a universal influenza vaccine.
Figures
References
-
- Assarsson, E., J. Sidney, C. Oseroff, V. Pasquetto, H. H. Bui, N. Frahm, C. Brander, B. Peters, H. Grey, and A. Sette. 2007. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J. Immunol. 1787890-7901. - PubMed
-
- Brown, D. M., A. M. Dilzer, D. L. Meents, and S. L. Swain. 2006. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J. Immunol. 1772888-2898. - PubMed
-
- Bui, H. H., J. Sidney, B. Peters, M. Sathiamurthy, A. Sinichi, K. A. Purton, B. R. Mothe, F. V. Chisari, D. I. Watkins, and A. Sette. 2005. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics 57304-314. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

