Analysis of copy number variants and segmental duplications in the human genome: Evidence for a change in the process of formation in recent evolutionary history
- PMID: 18842824
- PMCID: PMC2593581
- DOI: 10.1101/gr.081422.108
Analysis of copy number variants and segmental duplications in the human genome: Evidence for a change in the process of formation in recent evolutionary history
Abstract
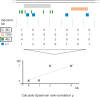
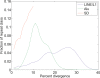
Segmental duplications (SDs) are operationally defined as >1 kb stretches of duplicated DNA with high sequence identity. They arise from copy number variants (CNVs) fixed in the population. To investigate the formation of SDs and CNVs, we examine their large-scale patterns of co-occurrence with different repeats. Alu elements, a major class of genomic repeats, had previously been identified as prime drivers of SD formation. We also observe this association; however, we find that it sharply decreases for younger SDs. Continuing this trend, we find only weak associations of CNVs with Alus. Similarly, we find an association of SDs with processed pseudogenes, which is decreasing for younger SDs and absent entirely for CNVs. Next, we find that SDs are significantly co-localized with each other, resulting in a highly skewed "power-law" distribution and chromosomal hotspots. We also observe a significant association of CNVs with SDs, but find that an SD-mediated mechanism only accounts for some CNVs (<28%). Overall, our results imply that a shift in predominant formation mechanism occurred in recent history: approximately 40 million years ago, during the "Alu burst" in retrotransposition activity, non-allelic homologous recombination, first mediated by Alus and then the by newly formed CNVs themselves, was the main driver of genome rearrangements; however, its relative importance has decreased markedly since then, with proportionally more events now stemming from other repeats and from non-homologous end-joining. In addition to a coarse-grained analysis, we performed targeted sequencing of 67 CNVs and then analyzed a combined set of 270 CNVs (540 breakpoints) to verify our conclusions.
Figures





References
-
- Albert R., Barabasi A.L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74:47–97.
-
- Bailey J.A., Eichler E.E. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. - PubMed
-
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. - PubMed
-
- Barabasi A.L., Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
