Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean
- PMID: 18842825
- PMCID: PMC2593655
- DOI: 10.1104/pp.108.127902
Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean
Abstract
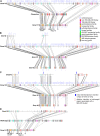
The genomes of most, if not all, flowering plants have undergone whole genome duplication events during their evolution. The impact of such polyploidy events is poorly understood, as is the fate of most duplicated genes. We sequenced an approximately 1 million-bp region in soybean (Glycine max) centered on the Rpg1-b disease resistance gene and compared this region with a region duplicated 10 to 14 million years ago. These two regions were also compared with homologous regions in several related legume species (a second soybean genotype, Glycine tomentella, Phaseolus vulgaris, and Medicago truncatula), which enabled us to determine how each of the duplicated regions (homoeologues) in soybean has changed following polyploidy. The biggest change was in retroelement content, with homoeologue 2 having expanded to 3-fold the size of homoeologue 1. Despite this accumulation of retroelements, over 77% of the duplicated low-copy genes have been retained in the same order and appear to be functional. This finding contrasts with recent analyses of the maize (Zea mays) genome, in which only about one-third of duplicated genes appear to have been retained over a similar time period. Fluorescent in situ hybridization revealed that the homoeologue 2 region is located very near a centromere. Thus, pericentromeric localization, per se, does not result in a high rate of gene inactivation, despite greatly accelerated retrotransposon accumulation. In contrast to low-copy genes, nucleotide-binding-leucine-rich repeat disease resistance gene clusters have undergone dramatic species/homoeologue-specific duplications and losses, with some evidence for partitioning of subfamilies between homoeologues.
Figures




References
-
- Akkaya MS, Bhagwat AA, Cregan PB (1995) Integration of simple sequence repeat DNA markers into a soybean linkage map. Crop Sci 35 1439–1445
-
- Ashfield T, Bocian A, Held D, Henk AD, Marek LF, Danesh D, Penuela S, Meksem K, Lightfoot DA, Young ND, et al (2003) Genetic and physical localization of the soybean Rpg1-b disease resistance gene reveals a complex locus containing several tightly linked families of NBS-LRR genes. Mol Plant Microbe Interact 16 817–826 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

