Posttranslational regulation of the scaffold for Fe-S cluster biogenesis, Isu
- PMID: 18843040
- PMCID: PMC2592649
- DOI: 10.1091/mbc.e08-06-0622
Posttranslational regulation of the scaffold for Fe-S cluster biogenesis, Isu
Abstract
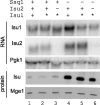
Isu, the scaffold protein on which Fe-S clusters are built in the mitochondrial matrix, plays a central role in the biogenesis of Fe-S cluster proteins. We report that the reduction in the activity of several components of the cluster biogenesis system, including the specialized Hsp70 Ssq1, causes a 15-20-fold up-regulation of Isu. This up-regulation results from changes at both the transcriptional and posttranslational level: an increase in ISU mRNA levels and in stability of ISU protein. Its biological importance is demonstrated by the fact that cells lacking Ssq1 grow poorly when Isu levels are prevented from rising above those found in wild-type cells. Of the biogenesis factors tested, Nfs1, the sulfur donor, was unique. Little increase in Isu levels occurred when Nfs1 was depleted. However, its presence was required for the up-regulation caused by reduction in activity of other components. Our results are consistent with the existence of a mechanism to increase the stability of Isu, and thus its level, that is dependent on the presence of the cysteine desulfurase Nfs1.
Figures






References
-
- Andrew A. J., Dutkiewicz R., Knieszner H., Craig E. A., Marszalek J. Characterization of the interaction between the J-protein Jac1p and the scaffold for Fe-S cluster biogenesis, Isu1p. J. Biol. Chem. 2006;281:14580–14587. - PubMed
-
- Ausubel F., Brent R., Kingston R., Moore D., Seidman J. G., Smith J., Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1997.
-
- Barras F., Loiseau L., Py B. How Escherichia coli and Saccharomyces cerevisiae build Fe/S proteins. Adv. Microb. Physiol. 2005;50:41–101. - PubMed
-
- Burns N., Grimwade B., Ross-Macdonald P. B., Choi E. Y., Finberg K., Roeder G. S., Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

