Remodeling of DNA replication structures by the branch point translocase FANCM
- PMID: 18843105
- PMCID: PMC2570989
- DOI: 10.1073/pnas.0804777105
Remodeling of DNA replication structures by the branch point translocase FANCM
Abstract
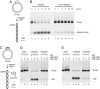
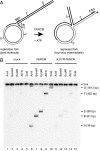
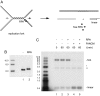
Fanconi anemia (FA) is a genetically heterogeneous chromosome instability syndrome associated with congenital abnormalities, bone marrow failure, and cancer predisposition. Eight FA proteins form a nuclear core complex, which promotes tolerance of DNA lesions in S phase, but the underlying mechanisms are still elusive. We reported recently that the FA core complex protein FANCM can translocate Holliday junctions. Here we show that FANCM promotes reversal of model replication forks via concerted displacement and annealing of the nascent and parental DNA strands. Fork reversal by FANCM also occurs when the lagging strand template is partially single-stranded and bound by RPA. The combined fork reversal and branch migration activities of FANCM lead to extensive regression of model replication forks. These observations provide evidence that FANCM can remodel replication fork structures and suggest a mechanism by which FANCM could promote DNA damage tolerance in S phase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks.Mol Cell. 2008 Jan 18;29(1):141-8. doi: 10.1016/j.molcel.2007.11.032. Mol Cell. 2008. PMID: 18206976
-
The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair.Mol Cell. 2008 Oct 10;32(1):118-28. doi: 10.1016/j.molcel.2008.08.024. Mol Cell. 2008. PMID: 18851838 Free PMC article.
-
The DNA translocase activity of FANCM protects stalled replication forks.Hum Mol Genet. 2012 May 1;21(9):2005-16. doi: 10.1093/hmg/dds013. Epub 2012 Jan 24. Hum Mol Genet. 2012. PMID: 22279085
-
A tumor suppressive DNA translocase named FANCM.Crit Rev Biochem Mol Biol. 2019 Feb;54(1):27-40. doi: 10.1080/10409238.2019.1568963. Epub 2019 Feb 4. Crit Rev Biochem Mol Biol. 2019. PMID: 30714416 Review.
-
Functions and regulation of the multitasking FANCM family of DNA motor proteins.Genes Dev. 2015 Sep 1;29(17):1777-88. doi: 10.1101/gad.266593.115. Genes Dev. 2015. PMID: 26341555 Free PMC article. Review.
Cited by
-
The ZGRF1 Helicase Promotes Recombinational Repair of Replication-Blocking DNA Damage in Human Cells.Cell Rep. 2020 Jul 7;32(1):107849. doi: 10.1016/j.celrep.2020.107849. Cell Rep. 2020. PMID: 32640219 Free PMC article.
-
The Simple Chordate Ciona intestinalis Has a Reduced Complement of Genes Associated with Fanconi Anemia.Evol Bioinform Online. 2016 Jun 6;12:133-48. doi: 10.4137/EBO.S37920. eCollection 2016. Evol Bioinform Online. 2016. PMID: 27279728 Free PMC article.
-
Antitumor efficacy of a sequence-specific DNA-targeted γPNA-based c-Myc inhibitor.Cell Rep Med. 2024 Jan 16;5(1):101354. doi: 10.1016/j.xcrm.2023.101354. Epub 2024 Jan 5. Cell Rep Med. 2024. PMID: 38183981 Free PMC article.
-
Expanded roles of the Fanconi anemia pathway in preserving genomic stability.Genes Dev. 2010 Aug 15;24(16):1680-94. doi: 10.1101/gad.1955310. Genes Dev. 2010. PMID: 20713514 Free PMC article. Review.
-
The Intra-S Checkpoint Responses to DNA Damage.Genes (Basel). 2017 Feb 17;8(2):74. doi: 10.3390/genes8020074. Genes (Basel). 2017. PMID: 28218681 Free PMC article. Review.
References
-
- Branzei D, Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amsterdam) 2007;6:994–1003. - PubMed
-
- Lambert S, Froget B, Carr AM. Arrested replication fork processing: Interplay between checkpoints and recombination. DNA Repair (Amsterdam) 2007;6:1042–1061. - PubMed
-
- Niedzwiedz W, et al. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. - PubMed
-
- Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2:446–457. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

