Generation of a highly inducible Gal4-->Fluc universal reporter mouse for in vivo bioluminescence imaging
- PMID: 18843112
- PMCID: PMC2572923
- DOI: 10.1073/pnas.0801075105
Generation of a highly inducible Gal4-->Fluc universal reporter mouse for in vivo bioluminescence imaging
Abstract
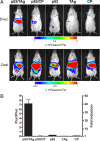
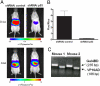
Full understanding of the functional complexity of the protein interactome requires mapping of biomolecular complexes within the cellular environment over biologically relevant time scales. New approaches to imaging interacting protein partners in vivo will allow the study of functional proteomics of human biology and disease within the context of living animals. Herein, we describe a universal transgenic reporter mouse strain that expresses firefly luciferase (Fluc) under the regulatory control of a concatenated Gal4 promoter (Tg(G4F(+/-))). Using an adenovirus to deliver a fused binding-domain-activator chimera (Gal4BD-VP16), induction of bioluminescence in Tg(G4F(+/-)) tissues of up to 4 orders of magnitude was observed in fibroblasts, liver, respiratory epithelia, muscle, and brain. The Tg(G4F(+/-)) reporter strain allowed noninvasive detection of viral infectivity, duration of the infection as well as viral clearance in various tissues in vivo. To demonstrate protein-protein interactions in live mice, the well characterized interaction between tumor suppressor p53 (fused to Gal4BD) and large T antigen (TAg) (fused to VP16) was visualized in vivo by using a two-hybrid strategy. Hepatocytes of Tg(G4F(+/-)) mice transfected with p53/TAg demonstrated 48-fold greater induction of Fluc expression in vivo than noninteracting pairs. Furthermore, to demonstrate the feasibility of monitoring experimental therapy with siRNA in vivo, targeted knockdown of p53 resulted in markedly reduced light output, whereas use of a control siRNA had no effect on protein interaction-dependent induction of Fluc. Thus, this highly inducible Gal4-->Fluc conditional reporter strain should facilitate imaging studies of protein interactions, signaling cascades, viral dissemination, and therapy within the physiological context of the whole animal.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Stark G, Kerr I, Williams B, Silverman R, Schreiber R. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. - PubMed
-
- Ogawa H, Ishiguro S, Gaubatz S, Livingston D, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. - PubMed
-
- Heldin C. Signal transduction: Multiple pathways, multiple options for therapy. Stem Cells. 2001;19:295–303. - PubMed
-
- Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

