The mitochondrial outer membrane protein hFis1 regulates mitochondrial morphology and fission through self-interaction
- PMID: 18845145
- PMCID: PMC2605612
- DOI: 10.1016/j.yexcr.2008.09.009
The mitochondrial outer membrane protein hFis1 regulates mitochondrial morphology and fission through self-interaction
Erratum in
- Exp Cell Res. 2009 Mar 10;315(5):899
Abstract
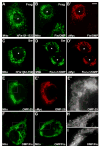
Mitochondrial fission in mammals is mediated by at least two proteins, DLP1/Drp1 and hFis1. DLP1 mediates the scission of mitochondrial membranes through GTP hydrolysis, and hFis1 is a putative DLP1 receptor anchored at the mitochondrial outer membrane by a C-terminal single transmembrane domain. The cytosolic domain of hFis1 contains six alpha-helices (alpha1-alpha6) out of which alpha2-alpha5 form two tetratricopeptide repeat (TPR) folds. In this study, by using chimeric constructs, we demonstrated that the cytosolic domain contains the necessary information for hFis1 function during mitochondrial fission. By using transient expression of different mutant forms of the hFis1 protein, we found that hFis1 self-interaction plays an important role in mitochondrial fission. Our results show that deletion of the alpha1 helix greatly increased the formation of dimeric and oligomeric forms of hFis1, indicating that alpha1 helix functions as a negative regulator of the hFis1 self-interaction. Further mutational approaches revealed that a tyrosine residue in the alpha5 helix and the linker between alpha3 and alpha4 helices participate in hFis1 oligomerization. Mutations causing oligomerization defect greatly reduced the ability to induce not only mitochondrial fragmentation by full-length hFis1 but also the formation of swollen ball-shaped mitochondria caused by alpha1-deleted hFis1. Our data suggest that oligomerization of hFis1 in the mitochondrial outer membrane plays a role in mitochondrial fission, potentially through participating in fission factor recruitment.
Figures







Similar articles
-
Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1.J Cell Sci. 2005 Sep 15;118(Pt 18):4141-51. doi: 10.1242/jcs.02537. Epub 2005 Aug 23. J Cell Sci. 2005. PMID: 16118244
-
Identification and characterization of unique proline-rich peptides binding to the mitochondrial fission protein hFis1.J Biol Chem. 2010 Jan 1;285(1):620-30. doi: 10.1074/jbc.M109.027508. Epub 2009 Oct 28. J Biol Chem. 2010. PMID: 19864424 Free PMC article.
-
The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1.Mol Cell Biol. 2003 Aug;23(15):5409-20. doi: 10.1128/MCB.23.15.5409-5420.2003. Mol Cell Biol. 2003. PMID: 12861026 Free PMC article.
-
New insights into the function and regulation of mitochondrial fission.Biochim Biophys Acta. 2013 May;1833(5):1256-68. doi: 10.1016/j.bbamcr.2013.02.002. Epub 2013 Feb 20. Biochim Biophys Acta. 2013. PMID: 23434681 Review.
-
[Advances in mitochondrial fusion-fission and Ca2+ signaling in mammals].Sheng Li Ke Xue Jin Zhan. 2010 Jun;41(3):171-6. Sheng Li Ke Xue Jin Zhan. 2010. PMID: 21416975 Review. Chinese.
Cited by
-
Control of mitochondrial morphology through differential interactions of mitochondrial fusion and fission proteins.PLoS One. 2011;6(5):e20655. doi: 10.1371/journal.pone.0020655. Epub 2011 May 27. PLoS One. 2011. PMID: 21647385 Free PMC article.
-
Homocysteine-mediated modulation of mitochondrial dynamics in retinal ganglion cells.Invest Ophthalmol Vis Sci. 2011 Jul 25;52(8):5551-8. doi: 10.1167/iovs.11-7256. Invest Ophthalmol Vis Sci. 2011. PMID: 21642619 Free PMC article.
-
Removal of a consensus proline is not sufficient to allow tetratricopeptide repeat oligomerization.Protein Sci. 2017 Oct;26(10):1974-1983. doi: 10.1002/pro.3234. Epub 2017 Jul 25. Protein Sci. 2017. PMID: 28707340 Free PMC article.
-
Mitochondrial fragmentation leads to intracellular acidification in Caenorhabditis elegans and mammalian cells.Mol Biol Cell. 2010 Jul 1;21(13):2191-201. doi: 10.1091/mbc.e09-10-0874. Epub 2010 May 5. Mol Biol Cell. 2010. PMID: 20444981 Free PMC article.
-
Origin of a folded repeat protein from an intrinsically disordered ancestor.Elife. 2016 Sep 13;5:e16761. doi: 10.7554/eLife.16761. Elife. 2016. PMID: 27623012 Free PMC article.
References
-
- Yoon Y. Regulation of mitochondrial dynamics: Another process modulated by Ca2+ signal? Sci STKE. 2005;2005:pe18. - PubMed
-
- Yoon Y. Sharpening the scissors: mitochondrial fission with aid. Cell Biochem Biophys. 2004;41:193–205. - PubMed
-
- Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–63. - PubMed
-
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, Bhattacharya SS, Wissinger B. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

