A simple and efficient expression and purification system using two newly constructed vectors
- PMID: 18845260
- PMCID: PMC3315830
- DOI: 10.1016/j.pep.2008.09.008
A simple and efficient expression and purification system using two newly constructed vectors
Abstract
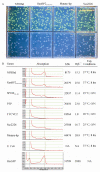
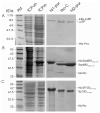
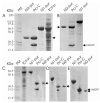
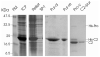
Structural biology places a high demand on proteins both in terms of quality and quantity. Although many protein expression and purification systems have been developed, an efficient and simple system which can be easily adapted is desirable. Here, we report a new system which combines improved expression, solubility screening and purification efficiency. The system is based on two newly constructed vectors, pEHISTEV and pEHISGFPTEV derived from a pET vector. Both vectors generate a construct with an amino-terminal hexahistidine tag (His-tag). In addition, pEHISGFPTEV expresses a protein with an N-terminal His-tagged green fluorescent protein (GFP) fusion to allow rapid quantitation of soluble protein. Both vectors have a tobacco etch virus (TEV) protease cleavage site that allows for production of protein with only two additional N-terminal residues and have the same multiple cloning site which enables parallel cloning. Protein purification is a simple two-stage nickel affinity chromatography based on the His tag removal. A total of seven genes were tested using this system. Expression was optimised using pEHISGFPTEV constructs by monitoring the GFP fluorescence and the soluble target proteins were quantified using spectrophotometric analysis. All the tested proteins were purified with sufficient quantity and quality to attempt structure determination. This system has been proven to be simple and effective for structural biology. The system is easily adapted to include other vectors, tags or fusions and therefore has the potential to be broadly applicable.
Figures



Similar articles
-
A family of E. coli expression vectors for laboratory scale and high throughput soluble protein production.BMC Biotechnol. 2006 Mar 1;6:12. doi: 10.1186/1472-6750-6-12. BMC Biotechnol. 2006. PMID: 16509985 Free PMC article.
-
Expression and Purification of Recombinant Proteins in Escherichia coli with a His6 or Dual His6-MBP Tag.Methods Mol Biol. 2017;1607:1-15. doi: 10.1007/978-1-4939-7000-1_1. Methods Mol Biol. 2017. PMID: 28573567 Free PMC article.
-
Plasmid system for the intracellular production and purification of affinity-tagged proteins in Bacillus megaterium.Biotechnol Bioeng. 2007 Feb 15;96(3):525-37. doi: 10.1002/bit.21145. Biotechnol Bioeng. 2007. PMID: 16964623
-
Simplifying Recombinant Protein Production: Combining Golden Gate Cloning with a Standardized Protein Purification Scheme.Methods Mol Biol. 2025;2850:229-249. doi: 10.1007/978-1-0716-4220-7_13. Methods Mol Biol. 2025. PMID: 39363075
-
In vivo cleavage of solubility tags as a tool to enhance the levels of soluble recombinant proteins in Escherichia coli.Biotechnol Bioeng. 2021 Nov;118(11):4159-4167. doi: 10.1002/bit.27912. Epub 2021 Aug 14. Biotechnol Bioeng. 2021. PMID: 34370304 Review.
Cited by
-
Development of a fluorescent monoclonal antibody-based assay to measure the allosteric effects of synthetic peptides on self-oligomerization of AGR2 protein.Protein Sci. 2013 Sep;22(9):1266-78. doi: 10.1002/pro.2299. Epub 2013 Jul 25. Protein Sci. 2013. PMID: 23780840 Free PMC article.
-
Cas6 specificity and CRISPR RNA loading in a complex CRISPR-Cas system.Nucleic Acids Res. 2014 Jun;42(10):6532-41. doi: 10.1093/nar/gku308. Epub 2014 Apr 20. Nucleic Acids Res. 2014. PMID: 24753403 Free PMC article.
-
Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut.Nat Microbiol. 2019 Dec;4(12):2393-2404. doi: 10.1038/s41564-019-0590-7. Epub 2019 Oct 21. Nat Microbiol. 2019. PMID: 31636419 Free PMC article.
-
Corrigendum: Pac13 is a Small Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.Angew Chem Int Ed Engl. 2020 Jul 27;59(31):12573. doi: 10.1002/anie.202000990. Angew Chem Int Ed Engl. 2020. PMID: 32691957 Free PMC article. No abstract available.
-
An improved strategy for easy process monitoring and advanced purification of recombinant proteins.Mol Biotechnol. 2013 Nov;55(3):227-35. doi: 10.1007/s12033-013-9673-5. Mol Biotechnol. 2013. PMID: 23780701
References
-
- Baum EZ, Bebernitz GA, Palant O, Mueller T, Plotch SJ. Purification, properties, and mutagenesis of poliovirus 3C protease. Virology. 1991;185:140–150. - PubMed
-
- Beis K, Srikannathasan V, Liu H, Fullerton SW, Bamford VA, Sanders DA, Whitfield C, Mcneil MR, Naismith JH. Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidised state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state. J. Mol. Biol. 2005;348:971–982. - PMC - PubMed
-
- Birch GM, Black T, Malcolm SK, Lai MT, Zimmerman RE, Jaskunas SR. Purification of recombinant human rhinovirus 14 3C protease expressed in Escherichia coli. Protein Expr. Purif. 1995;6:609–618. - PubMed
-
- Cabantous S, Pedelacq JD, Mark BL, Naranjo C, Terwilliger TC, Waldo GS. Recent advances in GFP folding reporter and split-GFP solubility reporter technologies. Application to improving the folding and solubility of recalcitrant proteins from Mycobacterium tuberculosis. J. Struct. Funct. Genomics. 2005;6:113–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

