The emerging role of splicing factors in cancer
- PMID: 18846105
- PMCID: PMC2581861
- DOI: 10.1038/embor.2008.189
The emerging role of splicing factors in cancer
Abstract
Recent progress in global sequence and microarray data analysis has revealed the increasing complexity of the human transcriptome. Alternative splicing generates a huge diversity of transcript variants and disruption of splicing regulatory networks is emerging as an important contributor to various diseases, including cancer. Current efforts to establish the dynamic repertoire of transcripts that are generated in health and disease are showing that many cancer-associated alternative-splicing events occur in the absence of mutations in the affected genes. A growing body of evidence reveals changes in splicing-factor expression that correlate with cancer development, progression and response to therapy. Here, we discuss how recent links between cancer and altered expression of proteins implicated in splicing regulation are bringing the splicing machinery to the fore as a potential target for anticancer treatment.
Figures
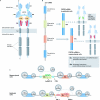

References
-
- Balasubramani M, Day BW, Schoen RE, Getzenberg RH (2006) Altered expression and localization of creatine kinase B, heterogeneous nuclear ribonucleoprotein F, and high mobility group box 1 protein in the nuclear matrix associated with colon cancer. Cancer Res 66: 763–769 - PubMed
-
- Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 - PubMed
-
- Brack SS, Silacci M, Birchler M, Neri D (2006) Tumor-targeting properties of novel antibodies specific to the large isoform of tenascin-C. Clin Cancer Res 12: 3200–3208 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

