Unique genes in plants: specificities and conserved features throughout evolution
- PMID: 18847470
- PMCID: PMC2576244
- DOI: 10.1186/1471-2148-8-280
Unique genes in plants: specificities and conserved features throughout evolution
Abstract
Background: Plant genomes contain a high proportion of duplicated genes as a result of numerous whole, segmental and local duplications. These duplications lead up to the formation of gene families, which are the usual material for many evolutionary studies. However, all characterized genomes include single-copy (unique) genes that have not received much attention. Unlike gene duplication, gene loss is not an unspecific mechanism but is rather influenced by a functional selection. In this context, we have established and used stringent criteria in order to identify suitable sets of unique genes present in plant proteomes. Comparisons of unique genes in the green phylum were used to characterize the gene and protein features exhibited by both conserved and species-specific unique genes.
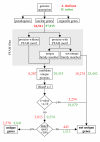
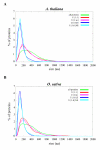
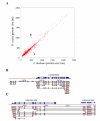
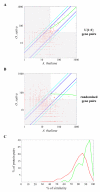
Results: We identified the unique genes within both A. thaliana and O. sativa genomes and classified them according to the number of homologs in the alternative species: none (U{1:0}), one (U{1:1}) or several (U{1:m}). Regardless of the species, all the genes in these groups present some conserved characteristics, such as small average protein size and abnormal intron number. In order to understand the origin and function of unique genes, we further characterized the U{1:1} gene pairs. The possible involvement of sequence convergence in the creation of U{1:1} pairs was discarded due to the frequent conservation of intron positions. Furthermore, an orthology relationship between the two members of each U{1:1} pair was strongly supported by a high conservation in the protein sizes and transcription levels. Within the promoter of the unique conserved genes, we found a number of TATA and TELO boxes that specifically differed from their mean number in the whole genome. Many unique genes have been conserved as unique through evolution from the green alga Ostreococcus lucimarinus to higher plants. Plant unique genes may also have homologs in bacteria and we showed a link between the targeting towards plastids of proteins encoded by plant nuclear unique genes and their homology with a bacterial protein.
Conclusion: Many of the A. thaliana and O. sativa unique genes are conserved in plants for which the ancestor diverged at least 725 million years ago (MYA). Half of these genes are also present in other eukaryotic and/or prokaryotic species. Thus, our results indicate that (i) a strong negative selection pressure has conserved a number of genes as unique in genomes throughout evolution, (ii) most unique genes are subjected to a low divergence rate, (iii) they have some features observed in housekeeping genes but for most of them there is no functional annotation and (iv) they may have an ancient origin involving a possible gene transfer from ancestral chloroplasts or bacteria to the plant nucleus.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

