SPSmart: adapting population based SNP genotype databases for fast and comprehensive web access
- PMID: 18847484
- PMCID: PMC2576268
- DOI: 10.1186/1471-2105-9-428
SPSmart: adapting population based SNP genotype databases for fast and comprehensive web access
Abstract
Background: In the last five years large online resources of human variability have appeared, notably HapMap, Perlegen and the CEPH foundation. These databases of genotypes with population information act as catalogues of human diversity, and are widely used as reference sources for population genetics studies. Although many useful conclusions may be extracted by querying databases individually, the lack of flexibility for combining data from within and between each database does not allow the calculation of key population variability statistics.
Results: We have developed a novel tool for accessing and combining large-scale genomic databases of single nucleotide polymorphisms (SNPs) in widespread use in human population genetics: SPSmart (SNPs for Population Studies). A fast pipeline creates and maintains a data mart from the most commonly accessed databases of genotypes containing population information: data is mined, summarized into the standard statistical reference indices, and stored into a relational database that currently handles as many as 4 x 10(9) genotypes and that can be easily extended to new database initiatives. We have also built a web interface to the data mart that allows the browsing of underlying data indexed by population and the combining of populations, allowing intuitive and straightforward comparison of population groups. All the information served is optimized for web display, and most of the computations are already pre-processed in the data mart to speed up the data browsing and any computational treatment requested.
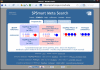
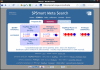
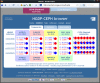
Conclusion: In practice, SPSmart allows populations to be combined into user-defined groups, while multiple databases can be accessed and compared in a few simple steps from a single query. It performs the queries rapidly and gives straightforward graphical summaries of SNP population variability through visual inspection of allele frequencies outlined in standard pie-chart format. In addition, full numerical description of the data is output in statistical results panels that include common population genetics metrics such as heterozygosity, Fst and In.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

