DNA repair of 8-oxo-7,8-dihydroguanine lesions in Porphyromonas gingivalis
- PMID: 18849425
- PMCID: PMC2593216
- DOI: 10.1128/JB.00919-08
DNA repair of 8-oxo-7,8-dihydroguanine lesions in Porphyromonas gingivalis
Abstract
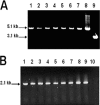
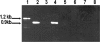
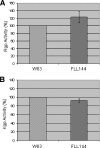
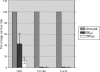
The persistence of Porphyromonas gingivalis in the inflammatory environment of the periodontal pocket requires an ability to overcome oxidative stress. DNA damage is a major consequence of oxidative stress. Unlike the case for other organisms, our previous report suggests a role for a non-base excision repair mechanism for the removal of 8-oxo-7,8-dihydroguanine (8-oxo-G) in P. gingivalis. Because the uvrB gene is known to be important in nucleotide excision repair, the role of this gene in the repair of oxidative stress-induced DNA damage was investigated. A 3.1-kb fragment containing the uvrB gene was PCR amplified from the chromosomal DNA of P. gingivalis W83. This gene was insertionally inactivated using the ermF-ermAM antibiotic cassette and used to create a uvrB-deficient mutant by allelic exchange. When plated on brucella blood agar, the mutant strain, designated P. gingivalis FLL144, was similar in black pigmentation and beta-hemolysis to the parent strain. In addition, P. gingivalis FLL144 demonstrated no significant difference in growth rate, proteolytic activity, or sensitivity to hydrogen peroxide from that of the parent strain. However, in contrast to the wild type, P. gingivalis FLL144 was significantly sensitive to UV irradiation. The enzymatic removal of 8-oxo-G from duplex DNA was unaffected by the inactivation of the uvrB gene. DNA affinity fractionation identified unique proteins that preferentially bound to the oligonucleotide fragment carrying the 8-oxo-G lesion. Collectively, these results suggest that the repair of oxidative stress-induced DNA damage involving 8-oxo-G may occur by a still undescribed mechanism in P. gingivalis.
Figures


Similar articles
-
Protective role of the PG1036-PG1037-PG1038 operon in oxidative stress in Porphyromonas gingivalis W83.PLoS One. 2013 Aug 19;8(8):e69645. doi: 10.1371/journal.pone.0069645. eCollection 2013. PLoS One. 2013. PMID: 23990885 Free PMC article.
-
Porphyromonas gingivalis mutY is involved in the repair of oxidative stress-induced DNA mispairing.Mol Oral Microbiol. 2011 Jun;26(3):175-86. doi: 10.1111/j.2041-1014.2011.00605.x. Epub 2011 Feb 22. Mol Oral Microbiol. 2011. PMID: 21545695 Free PMC article.
-
The bcp gene in the bcp-recA-vimA-vimE-vimF operon is important in oxidative stress resistance in Porphyromonas gingivalis W83.Mol Oral Microbiol. 2011 Feb;26(1):62-77. doi: 10.1111/j.2041-1014.2010.00596.x. Epub 2010 Nov 18. Mol Oral Microbiol. 2011. PMID: 21214873 Free PMC article.
-
Gingipain-dependent interactions with the host are important for survival of Porphyromonas gingivalis.Front Biosci. 2008 May 1;13:3215-38. doi: 10.2741/2922. Front Biosci. 2008. PMID: 18508429 Free PMC article. Review.
-
VimA mediates multiple functions that control virulence in Porphyromonas gingivalis.Mol Oral Microbiol. 2013 Jun;28(3):167-80. doi: 10.1111/omi.12017. Epub 2012 Dec 21. Mol Oral Microbiol. 2013. PMID: 23279905 Free PMC article. Review.
Cited by
-
Oxidative stress resistance in Porphyromonas gingivalis.Future Microbiol. 2012 Apr;7(4):497-512. doi: 10.2217/fmb.12.17. Future Microbiol. 2012. PMID: 22439726 Free PMC article. Review.
-
Photoinactivation and Photoablation of Porphyromonas gingivalis.Pathogens. 2023 Sep 14;12(9):1160. doi: 10.3390/pathogens12091160. Pathogens. 2023. PMID: 37764967 Free PMC article. Review.
-
Nitric oxide stress resistance in Porphyromonas gingivalis is mediated by a putative hydroxylamine reductase.J Bacteriol. 2012 Mar;194(6):1582-92. doi: 10.1128/JB.06457-11. Epub 2012 Jan 13. J Bacteriol. 2012. PMID: 22247513 Free PMC article.
-
Protective role of the PG1036-PG1037-PG1038 operon in oxidative stress in Porphyromonas gingivalis W83.PLoS One. 2013 Aug 19;8(8):e69645. doi: 10.1371/journal.pone.0069645. eCollection 2013. PLoS One. 2013. PMID: 23990885 Free PMC article.
-
VimA-dependent modulation of acetyl coenzyme A levels and lipid A biosynthesis can alter virulence in Porphyromonas gingivalis.Infect Immun. 2012 Feb;80(2):550-64. doi: 10.1128/IAI.06062-11. Epub 2011 Dec 5. Infect Immun. 2012. PMID: 22144476 Free PMC article.
References
-
- Abaibou, H., Q. Ma, G. J. Olango, J. Potempa, J. Travis, and H. M. Fletcher. 2000. Unaltered expression of the major protease genes in a non-virulent recA-defective mutant of Porphyromonas gingivalis W83. Oral Microbiol. Immunol. 1540-47. - PubMed
-
- Alleva, J. L., S. Zuo, J. Hurwitz, and P. W. Doetsch. 2000. In vitro reconstitution of the Schizosaccharomyces pombe alternative excision repair pathway. Biochemistry 392659-2666. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

