Stem-loop silencing reveals that a third mitochondrial DNA polymerase, POLID, is required for kinetoplast DNA replication in trypanosomes
- PMID: 18849470
- PMCID: PMC2593185
- DOI: 10.1128/EC.00199-08
Stem-loop silencing reveals that a third mitochondrial DNA polymerase, POLID, is required for kinetoplast DNA replication in trypanosomes
Abstract
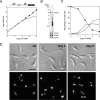
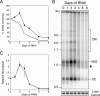
Kinetoplast DNA (kDNA), the mitochondrial genome of trypanosomes, is a catenated network containing thousands of minicircles and tens of maxicircles. The topological complexity dictates some unusual features including a topoisomerase-mediated release-and-reattachment mechanism for minicircle replication and at least six mitochondrial DNA polymerases (Pols) for kDNA transactions. Previously, we identified four family A DNA Pols from Trypanosoma brucei with similarity to bacterial DNA Pol I and demonstrated that two (POLIB and POLIC) were essential for maintaining the kDNA network, while POLIA was not. Here, we used RNA interference to investigate the function of POLID in procyclic T. brucei. Stem-loop silencing of POLID resulted in growth arrest and the progressive loss of the kDNA network. Additional defects in kDNA replication included a rapid decline in minicircle and maxicircle abundance and a transient accumulation of minicircle replication intermediates before loss of the kDNA network. These results demonstrate that POLID is a third essential DNA Pol required for kDNA replication. While other eukaryotes utilize a single DNA Pol (Pol gamma) for replication of mitochondrial DNA, T. brucei requires at least three to maintain the complex kDNA network.
Figures
Similar articles
-
A DNA polymerization-independent role for mitochondrial DNA polymerase I-like protein C in African trypanosomes.J Cell Sci. 2020 May 7;133(9):jcs233072. doi: 10.1242/jcs.233072. J Cell Sci. 2020. PMID: 32079654 Free PMC article.
-
Mitochondrial DNA polymerase POLIB is essential for minicircle DNA replication in African trypanosomes.Mol Microbiol. 2010 Mar;75(6):1414-25. doi: 10.1111/j.1365-2958.2010.07061.x. Epub 2009 Feb 1. Mol Microbiol. 2010. PMID: 20132449
-
Multiple mitochondrial DNA polymerases in Trypanosoma brucei.Mol Cell. 2002 Jul;10(1):175-86. doi: 10.1016/s1097-2765(02)00571-3. Mol Cell. 2002. PMID: 12150917
-
Mitochondrial genome maintenance-the kinetoplast story.FEMS Microbiol Rev. 2023 Nov 1;47(6):fuac047. doi: 10.1093/femsre/fuac047. FEMS Microbiol Rev. 2023. PMID: 36449697 Free PMC article. Review.
-
The structure and replication of kinetoplast DNA.Curr Mol Med. 2004 Sep;4(6):623-47. doi: 10.2174/1566524043360096. Curr Mol Med. 2004. PMID: 15357213 Review.
Cited by
-
Three mitochondrial DNA polymerases are essential for kinetoplast DNA replication and survival of bloodstream form Trypanosoma brucei.Eukaryot Cell. 2011 Jun;10(6):734-43. doi: 10.1128/EC.05008-11. Epub 2011 Apr 29. Eukaryot Cell. 2011. PMID: 21531873 Free PMC article.
-
Expression, purification, and biochemical characterization of recombinant DNA polymerase beta of the Trypanosoma cruzi TcI lineage: requirement of additional factors and detection of phosphorylation of the native form.Parasitol Res. 2015 Apr;114(4):1313-26. doi: 10.1007/s00436-014-4308-8. Epub 2015 Jan 9. Parasitol Res. 2015. PMID: 25566774
-
A DNA polymerization-independent role for mitochondrial DNA polymerase I-like protein C in African trypanosomes.J Cell Sci. 2020 May 7;133(9):jcs233072. doi: 10.1242/jcs.233072. J Cell Sci. 2020. PMID: 32079654 Free PMC article.
-
Mitochondrial heat shock protein machinery hsp70/hsp40 is indispensable for proper mitochondrial DNA maintenance and replication.mBio. 2015 Feb 10;6(1):e02425-14. doi: 10.1128/mBio.02425-14. mBio. 2015. PMID: 25670781 Free PMC article.
-
Knockdown of Inner Arm Protein IC138 in Trypanosoma brucei Causes Defective Motility and Flagellar Detachment.PLoS One. 2015 Nov 10;10(11):e0139579. doi: 10.1371/journal.pone.0139579. eCollection 2015. PLoS One. 2015. PMID: 26555902 Free PMC article.
References
-
- Brun, R., and Schonenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36289-292. - PubMed
-
- Chanez, A. L., A. B. Hehl, M. Engstler, and A. Schneider. 2006. Ablation of the single dynamin of T. brucei blocks mitochondrial fission and endocytosis and leads to a precise cytokinesis arrest. J. Cell Sci. 1192968-2974. - PubMed
-
- Chen, Y., C. H. Hung, T. Burderer, and G. S. Lee. 2003. Development of RNA interference revertants in Trypanosoma brucei cell lines generated with a double stranded RNA expression construct driven by two opposing promoters. Mol. Biochem. Parasitol. 126275-279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

