Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis
- PMID: 18849490
- PMCID: PMC2590739
- DOI: 10.1105/tpc.108.062760
Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis
Abstract
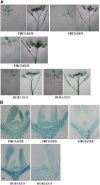
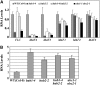
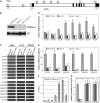
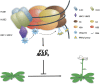
Ubiquitination is one of many known histone modifications that regulate gene expression. Here, we examine the Arabidopsis thaliana homologs of the yeast E2 and E3 enzymes responsible for H2B monoubiquitination (H2Bub1). Arabidopsis has two E3 homologs (HISTONE MONOUBIQUITINATION1 [HUB1] and HUB2) and three E2 homologs (UBIQUITIN CARRIER PROTEIN [UBC1] to UBC3). hub1 and hub2 mutants show the loss of H2Bub1 and early flowering. By contrast, single ubc1, ubc2, or ubc3 mutants show no flowering defect; only ubc1 ubc2 double mutants, and not double mutants with ubc3, show early flowering and H2Bub1 defects. This suggests that ubc1 and ubc2 are redundant, but ubc3 is not involved in flowering time regulation. Protein interaction analysis showed that HUB1 and HUB2 interact with each other and with UBC1 and UBC2, as well as self-associating. The expression of FLOWERING LOCUS C (FLC) and its homologs was repressed in hub1, hub2, and ubc1 ubc2 mutant plants. Association of H2Bub1 with the chromatin of FLC clade genes depended on UBC1,2 and HUB1,2, as did the dynamics of methylated histones H3K4me3 and H3K36me2. The monoubiquitination of H2B via UBC1,2 and HUB1,2 represents a novel form of histone modification that is involved in flowering time regulation.
Figures







References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. - PubMed
-
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methlation. Nature 427 164–167. - PubMed
-
- Baurle, I., and Dean, C. (2006). The timing of developmental transition in plants. Cell 125 655–664. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

