In-depth characterization of the microRNA transcriptome in a leukemia progression model
- PMID: 18849523
- PMCID: PMC2577858
- DOI: 10.1101/gr.077578.108
In-depth characterization of the microRNA transcriptome in a leukemia progression model
Abstract
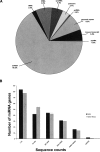
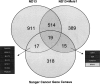
MicroRNAs (miRNAs) have been shown to play important roles in physiological as well as multiple malignant processes, including acute myeloid leukemia (AML). In an effort to gain further insight into the role of miRNAs in AML, we have applied the Illumina massively parallel sequencing platform to carry out an in-depth analysis of the miRNA transcriptome in a murine leukemia progression model. This model simulates the stepwise conversion of a myeloid progenitor cell by an engineered overexpression of the nucleoporin 98 (NUP98)-homeobox HOXD13 fusion gene (ND13), to aggressive AML inducing cells upon transduction with the oncogenic collaborator Meis1. From this data set, we identified 307 miRNA/miRNA species in the ND13 cells and 306 miRNA/miRNA species in ND13+Meis1 cells, corresponding to 223 and 219 miRNA genes. Sequence counts varied between two and 136,558, indicating a remarkable expression range between the detected miRNA species. The large number of miRNAs expressed and the nature of differential expression suggest that leukemic progression as modeled here is dictated by the repertoire of shared, but differentially expressed miRNAs. Our finding of extensive sequence variations (isomiRs) for almost all miRNA and miRNA species adds additional complexity to the miRNA transcriptome. A stringent target prediction analysis coupled with in vitro target validation revealed the potential for miRNA-mediated release of oncogenes that facilitates leukemic progression from the preleukemic to leukemia inducing state. Finally, 55 novel miRNAs species were identified in our data set, adding further complexity to the emerging world of small RNAs.
Figures


References
-
- Akao Y., Nakagawa Y., Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006;29:903–906. - PubMed
-
- Aravin A., Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–5840. - PubMed
-
- Argiropoulos B., Palmqvist L., Yung E., Kuchenbauer F., Heuser M., Sly L.M., Wan A., Krystal G., Humphries R.K. Linkage of Meis1 leukemogenic activity to multiple downstream effectors including Trib2 and Ccl3. Exp. Hematol. 2008;36:845–859. - PubMed
-
- Bandres E., Agirre X., Ramirez N., Zarate R., Garcia-Foncillas J. MicroRNAs as cancer players: Potential clinical and biological effects. DNA Cell Biol. 2007;26:273–282. - PubMed
-
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
