HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot
- PMID: 18849571
- PMCID: PMC2686602
- DOI: 10.1093/nar/gkn661
HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot
Abstract
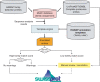
The growth in the number of completely sequenced microbial genomes (bacterial and archaeal) has generated a need for a procedure that provides UniProtKB/Swiss-Prot-quality annotation to as many protein sequences as possible. We have devised a semi-automated system, HAMAP (High-quality Automated and Manual Annotation of microbial Proteomes), that uses manually built annotation templates for protein families to propagate annotation to all members of manually defined protein families, using very strict criteria. The HAMAP system is composed of two databases, the proteome database and the family database, and of an automatic annotation pipeline. The proteome database comprises biological and sequence information for each completely sequenced microbial proteome, and it offers several tools for CDS searches, BLAST options and retrieval of specific sets of proteins. The family database currently comprises more than 1500 manually curated protein families and their annotation templates that are used to annotate proteins that belong to one of the HAMAP families. On the HAMAP website, individual sequences as well as whole genomes can be scanned against all HAMAP families. The system provides warnings for the absence of conserved amino acid residues, unusual sequence length, etc. Thanks to the implementation of HAMAP, more than 200,000 microbial proteins have been fully annotated in UniProtKB/Swiss-Prot (HAMAP website: http://www.expasy.org/sprot/hamap).
Figures


References
-
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb J.-F, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. - PubMed
-
- Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. - PubMed
-
- Stothard P, Wishart DS. Automated bacterial genome analysis and annotation. Curr. Opin. Microbiol. 2006;9:505–510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

