DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes
- PMID: 18849970
- PMCID: PMC2605662
- DOI: 10.1038/nature07392
DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes
Abstract
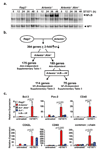
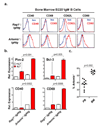
DNA double-strand breaks are generated by genotoxic agents and by cellular endonucleases as intermediates of several important physiological processes. The cellular response to genotoxic DNA breaks includes the activation of transcriptional programs known primarily to regulate cell-cycle checkpoints and cell survival. DNA double-strand breaks are generated in all developing lymphocytes during the assembly of antigen receptor genes, a process that is essential for normal lymphocyte development. Here we show that in murine lymphocytes these physiological DNA breaks activate a broad transcriptional program. This program transcends the canonical DNA double-strand break response and includes many genes that regulate diverse cellular processes important for lymphocyte development. Moreover, the expression of several of these genes is regulated similarly in response to genotoxic DNA damage. Thus, physiological DNA double-strand breaks provide cues that can regulate cell-type-specific processes not directly involved in maintaining the integrity of the genome, and genotoxic DNA breaks could disrupt normal cellular functions by corrupting these processes.
Figures


References
-
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3(3):155–168. - PubMed
-
- Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297(5581):547–551. - PubMed
-
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408(6811):433–439. - PubMed
-
- Innes CL, et al. ATM requirement in gene expression responses to ionizing radiation in human lymphoblasts and fibroblasts. Mol Cancer Res. 2006;4(3):197–207. - PubMed
-
- Rashi-Elkeles S, et al. Parallel induction of ATM-dependent pro- and antiapoptotic signals in response to ionizing radiation in murine lymphoid tissue. Oncogene. 2006;25(10):1584–1592. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

