A simple, fast, and accurate method of phylogenomic inference
- PMID: 18851752
- PMCID: PMC2760878
- DOI: 10.1186/gb-2008-9-10-r151
A simple, fast, and accurate method of phylogenomic inference
Abstract
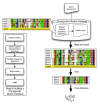
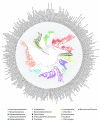
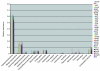
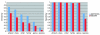
The explosive growth of genomic data provides an opportunity to make increased use of protein markers for phylogenetic inference. We have developed an automated pipeline for phylogenomic analysis (AMPHORA) that overcomes the existing bottlenecks limiting large-scale protein phylogenetic inference. We demonstrated its high throughput capabilities and high quality results by constructing a genome tree of 578 bacterial species and by assigning phylotypes to 18,607 protein markers identified in metagenomic data collected from the Sargasso Sea.
Figures
References
-
- Woese CR, Achenbach L, Rouviere P, Mandelco L. Archaeal phylogeny: reexamination of the phylogenetic position of Archaeoglobus fulgidus in light of certain composition-induced artifacts. Syst Appl Microbiol. 1991;14:364–371. - PubMed
-
- Ludwig W, Klenk H-P. Overview: A phylogenetic backbone and taxonomic framework for procaryotic systematics. In: Boone DR, Castenholz RW, Garrity GM, editor. Bergey's Manual of Systematic Bacteriology. 2. Vol. 1. New York, NY: Springer-Verlag; 2000. pp. 49–65.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

