The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair
- PMID: 18851838
- PMCID: PMC2581491
- DOI: 10.1016/j.molcel.2008.08.024
The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair
Abstract
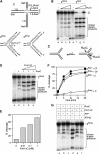
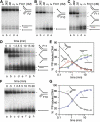
The Fanconi anemia (FA) core complex promotes the tolerance/repair of DNA damage at stalled replication forks by catalyzing the monoubiquitination of FANCD2 and FANCI. Intriguingly, the core complex component FANCM also catalyzes branch migration of model Holliday junctions and replication forks in vitro. Here we have characterized the ortholog of FANCM in fission yeast Fml1 in order to understand the physiological significance of this activity. We show that Fml1 has at least two roles in homologous recombination-it promotes Rad51-dependent gene conversion at stalled/blocked replication forks and limits crossing over during mitotic double-strand break repair. In vitro Fml1 catalyzes both replication fork reversal and D loop disruption, indicating possible mechanisms by which it can fulfill its pro- and antirecombinogenic roles.
Figures

References
-
- Bugreev D.V., Mazina O.M., Mazin A.V. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

