Stat3 contributes to resistance toward BCR-ABL inhibitors in a bone marrow microenvironment model of drug resistance
- PMID: 18852120
- PMCID: PMC2676735
- DOI: 10.1158/1535-7163.MCT-08-0314
Stat3 contributes to resistance toward BCR-ABL inhibitors in a bone marrow microenvironment model of drug resistance
Abstract
Imatinib mesylate is a potent, molecularly targeted therapy against the oncogenic tyrosine kinase BCR-ABL. Although imatinib mesylate has considerable efficacy against chronic myeloid leukemia (CML), advanced-stage CML patients frequently become refractory to this agent. The bone marrow is the predominant microenvironment of CML and is a rich source of both soluble factors and extracellular matrices, which may influence drug response. To address the influence of the bone marrow microenvironment on imatinib mesylate sensitivity, we used an in vitro bone marrow stroma model. Our data show culturing K562 cells, in bone marrow stroma-derived conditioned medium (CM), is sufficient to cause resistance to BCR-ABL inhibitors. Drug resistance correlated with increased pTyrStat3, whereas no increases in pTyrStat5 was noted. Moreover, resistance was associated with increased levels of the Stat3 target genes Bcl-xl, Mcl-1, and survivin. Finally, reducing Stat3 levels with small interfering RNA sensitized K562 cells cultured in CM to imatinib mesylate-induced cell death. Importantly, Stat3 dependency was specific for cells grown in CM, as reducing Stat3 levels in regular growth conditions had no effect on imatinib mesylate sensitivity. Together, these data support a novel mechanism of BCR-ABL-independent imatinib mesylate resistance and provides preclinical rationale for using Stat3-inhibitors to increase the efficacy of imatinib mesylate within the context of the bone marrow microenvironment.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures
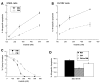
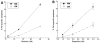
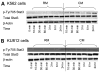



References
-
- Bartram CR, de Klein A, Hagemeijer A, et al. Translocation of c-ab1 oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1983;306:277–80. - PubMed
-
- Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–9. - PubMed
-
- Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247:1079–82. - PubMed
-
- Buchdunger E, Zimmermann J, Mett H, et al. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996;56:100–4. - PubMed
-
- Druker BJ, Guilhot F, O’Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

