Development and qualification of a pharmacodynamic model for the pronounced inoculum effect of ceftazidime against Pseudomonas aeruginosa
- PMID: 18852268
- PMCID: PMC2612150
- DOI: 10.1128/AAC.00489-08
Development and qualification of a pharmacodynamic model for the pronounced inoculum effect of ceftazidime against Pseudomonas aeruginosa
Abstract
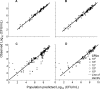
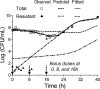
Evidence is mounting in support of the inoculum effect (i.e., slow killing at large initial inocula [CFUo]) for numerous antimicrobials against a variety of pathogens. Our objectives were to (i) determine the impact of the CFUo of Pseudomonas aeruginosa on ceftazidime activity and (ii) to develop and validate a pharmacokinetic/pharmacodynamic (PKPD) mathematical model accommodating a range of CFUo. Time-kill experiments using ceftazidime at seven concentrations up to 128 mg/liter (MIC, 2 mg/liter) were performed in duplicate against P. aeruginosa PAO1 at five CFUo from 10(5) to 10(9) CFU/ml. Samples were collected over 24 h and fit by candidate models in NONMEM VI and S-ADAPT 1.55 (all data were comodeled). External model qualification integrated data from eight previously published studies. Ceftazidime displayed approximately 3 to 4 log(10) CFU/ml net killing at 10(6.2) CFUo and concentrations of 4 mg/liter (or higher), less than 1.6 log(10) CFU/ml killing at 10(7.3) CFUo, and no killing at 10(8.0) CFUo for concentrations up to 128 mg/liter. The proposed mechanism-based model successfully described the inoculum effect and the concentration-independent lag time of killing. The mean generation time was 28.3 min. The effect of an autolysin was assumed to inhibit successful replication. Ceftazidime concentrations of 0.294 mg/liter stimulated the autolysin effect by 50%. The model was predictive in the internal cross-validation and had excellent in silico predictive performance for published studies of P. aeruginosa ATCC 27853 for various CFUo. The proposed PKPD model successfully described and predicted the pronounced inoculum effect of ceftazidime in vitro and integrated data from eight literature studies to support translation from time-kill experiments to in vitro infection models.
Figures





References
-
- Barclay, M. L., E. J. Begg, S. T. Chambers, and B. A. Peddie. 1996. The effect of aminoglycoside-induced adaptive resistance on the antibacterial activity of other antibiotics against Pseudomonas aeruginosa in vitro. J. Antimicrob. Chemother. 38:853-858. - PubMed
-
- Bauer, R. J. 2007. S-ADAPT/MCPEM user's guide (version 1.55). Pharmacokinetic, Pharmacodynamic and Population Data Analysis, Berkeley, CA.
-
- Beal, S. L., L. B. Sheiner, and A. J. Boeckmann. 2006. NONMEM users guides (1989-2006). Icon Development Solutions, Ellicott City, MD.
-
- Bratu, S., J. Gupta, and J. Quale. 2006. Expression of the las and rhl quorum-sensing systems in clinical isolates of Pseudomonas aeruginosa does not correlate with efflux pump expression or antimicrobial resistance. J. Antimicrob. Chemother. 58:1250-1253. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

