Detection of staphylococcal cassette chromosome mec-associated DNA segments in multiresistant methicillin-susceptible Staphylococcus aureus (MSSA) and identification of Staphylococcus epidermidis ccrAB4 in both methicillin-resistant S. aureus and MSSA
- PMID: 18852274
- PMCID: PMC2592854
- DOI: 10.1128/AAC.00447-08
Detection of staphylococcal cassette chromosome mec-associated DNA segments in multiresistant methicillin-susceptible Staphylococcus aureus (MSSA) and identification of Staphylococcus epidermidis ccrAB4 in both methicillin-resistant S. aureus and MSSA
Abstract
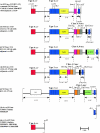
Methicillin-susceptible Staphylococcus aureus (MSSA) can arise from methicillin-resistant S. aureus (MRSA) following partial or complete excision of staphylococcal cassette chromosome mec (SCCmec). This study investigated whether multiresistant MSSA isolates from Irish hospitals, where MRSA has been endemic for decades, harbor SCCmec DNA. Twenty-five multiresistant MSSA isolates recovered between 2002 and 2006 were tested for SCCmec DNA by PCR and were genotyped by multilocus sequence typing and spa typing. All isolates lacked mecA. Three isolates (12%) harbored SCCmec DNA; two of these (genotype ST8/t190) harbored a 26-kb SCCmec IID (II.3.1.2) remnant that lacked part of mecI and all of mecR1, mecA, and IS431; the third isolate (ST8/t3209) harbored the SCCmec region from dcs to orfX. All three isolates were detected as MRSA using the BD GeneOhm and Cepheid's Xpert MRSA real-time PCR assays. Six isolates (ST8/t190, n = 4; ST5/t088, n = 2), including both isolates with the SCCmec IID remnant, harbored ccrAB4 with 100% identity to ccrAB4 from the Staphylococcus epidermidis composite island SCC-CI. This ccrAB4 gene was also identified in 23 MRSA isolates representative of ST8/t190-MRSA with variant SCCmec II subtypes IIA to IIE, which predominated previously in Irish hospitals. ccrAB4 was located 5,549 bp upstream of the left SCCmec junction in both the MRSA and MSSA isolates with SCCmec elements and remnants and 5,549 bp upstream of orfX in the four MSSA isolates with ccrAB4 only on an SCC-CI homologous region. This is the first description of a large SCCmec remnant with ccr and partial mec genes in MSSA and of the S. epidermidis SCC-CI and ccrAB4 genes in S. aureus.
Figures
References
-
- Abbott, Y., N. Leonard, B. Markey, A. Rossney, and J. Dunne. 2007. Persistence of MRSA infection. Vet. Rec. 160:851-852. - PubMed
-
- Carroll, D., M. A. Kehoe, D. Cavanagh, and D. C. Coleman. 1995. Novel organization of the site-specific integration and excision recombination functions of the Staphylococcus aureus serotype F virulence-converting phages phi 13 and phi 42. Mol. Microbiol. 16:877-893. - PubMed
-
- Collery, M. M., D. S. Smyth, J. M. Twohig, A. C. Shore, D. C. Coleman, and C. J. Smyth. 2008. Molecular typing of nasal carriage isolates of Staphylococcus aureus from an Irish university student population based on toxin gene PCR, agr locus types and multiple locus, variable number tandem repeat analysis. J. Med. Microbiol. 57:348-358. - PubMed
-
- Cookson, B. D., D. A. Robinson, A. B. Monk, S. Murchan, A. Deplano, R. de Ryck, M. J. Struelens, C. Scheel, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, C. Cuny, W. Witte, P. T. Tassios, N. J. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, M. Muller-Premru, W. Hryniewicz, A. Rossney, B. O'Connell, B. D. Short, J. Thomas, S. O'Hanlon, and M. C. Enright. 2007. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. J. Clin. Microbiol. 45:1830-1837. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

