MicroRNA-directed transcriptional gene silencing in mammalian cells
- PMID: 18852463
- PMCID: PMC2571020
- DOI: 10.1073/pnas.0808830105
MicroRNA-directed transcriptional gene silencing in mammalian cells
Abstract
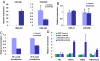
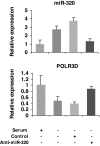
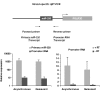
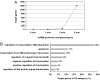
MicroRNAs (miRNAs) regulate gene expression at the posttranscriptional level in the cytoplasm, but recent findings suggest additional roles for miRNAs in the nucleus. To address whether miRNAs might transcriptionally silence gene expression, we searched for miRNA target sites proximal to known gene transcription start sites in the human genome. One conserved miRNA, miR-320, is encoded within the promoter region of the cell cycle gene POLR3D in the antisense orientation. We provide evidence of a cis-regulatory role for miR-320 in transcriptional silencing of POLR3D expression. miR-320 directs the association of RNA interference (RNAi) protein Argonaute-1 (AGO1), Polycomb group (PcG) component EZH2, and tri-methyl histone H3 lysine 27 (H3K27me3) with the POLR3D promoter. Our results suggest the existence of an epigenetic mechanism of miRNA-directed transcriptional gene silencing (TGS) in mammalian cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Zaratiegui M, et al. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. - PubMed
-
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. - PubMed
-
- Kim DH, et al. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

