Accuracy of predicting the genetic risk of disease using a genome-wide approach
- PMID: 18852893
- PMCID: PMC2561058
- DOI: 10.1371/journal.pone.0003395
Accuracy of predicting the genetic risk of disease using a genome-wide approach
Abstract
Background: The prediction of the genetic disease risk of an individual is a powerful public health tool. While predicting risk has been successful in diseases which follow simple Mendelian inheritance, it has proven challenging in complex diseases for which a large number of loci contribute to the genetic variance. The large numbers of single nucleotide polymorphisms now available provide new opportunities for predicting genetic risk of complex diseases with high accuracy.
Methodology/principal findings: We have derived simple deterministic formulae to predict the accuracy of predicted genetic risk from population or case control studies using a genome-wide approach and assuming a dichotomous disease phenotype with an underlying continuous liability. We show that the prediction equations are special cases of the more general problem of predicting the accuracy of estimates of genetic values of a continuous phenotype. Our predictive equations are responsive to all parameters that affect accuracy and they are independent of allele frequency and effect distributions. Deterministic prediction errors when tested by simulation were generally small. The common link among the expressions for accuracy is that they are best summarized as the product of the ratio of number of phenotypic records per number of risk loci and the observed heritability.
Conclusions/significance: This study advances the understanding of the relative power of case control and population studies of disease. The predictions represent an upper bound of accuracy which may be achievable with improved effect estimation methods. The formulae derived will help researchers determine an appropriate sample size to attain a certain accuracy when predicting genetic risk.
Conflict of interest statement
Figures
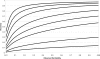
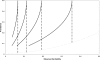
 , so values below this line are not possible under the liability model.
, so values below this line are not possible under the liability model.References
-
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nature Genetics. 2006;38:879–887. - PubMed
-
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Harlow, UK: Longman; 1996.
-
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nature Reviews Genetics. 2005;6:95–108. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

