Wagner and Dollo: a stochastic duet by composing two parsimonious solos
- PMID: 18853363
- PMCID: PMC4677801
- DOI: 10.1080/10635150802434394
Wagner and Dollo: a stochastic duet by composing two parsimonious solos
Abstract
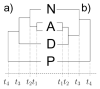
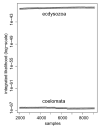
New contributions toward generalizing evolutionary models expand greatly our ability to analyze complex evolutionary characters and advance phylogeny reconstruction. In this article, we extend the binary stochastic Dollo model to allow for multi-state characters. In doing so, we align previously incompatible Wagner and Dollo parsimony principles under a common probabilistic framework by embedding arbitrary continuous-time Markov chains into the binary stochastic Dollo model. This approach enables us to analyze character traits that exhibit both Dollo and Wagner characteristics throughout their evolutionary histories. Utilizing Bayesian inference, we apply our novel model to analyze intron conservation patterns and the evolution of alternatively spliced exons. The generalized framework we develop demonstrates potential in distinguishing between phylogenetic hypotheses and providing robust estimates of evolutionary rates. Moreover, for the two applications analyzed here, our framework is the first to provide an adequate stochastic process for the data. We discuss possible extensions to the framework from both theoretical and applied perspectives.
Figures

References
-
- Aguinaldo AMA, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. - PubMed
-
- Brooks SP, Gelman A. General methods for monitoring convergence of iterative simulations. J. Comput. Graph. Stat. 1998;7:434–455.
-
- Cavender JA, Felsenstein J. Invariants of phylogenies in a simple case with discrete states. J. Classif. 1987;4:57–71.
-
- Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

