Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication
- PMID: 18854154
- PMCID: PMC2628946
- DOI: 10.1016/j.cell.2008.07.032
Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication
Abstract
Human Immunodeficiency Viruses (HIV-1 and HIV-2) rely upon host-encoded proteins to facilitate their replication. Here, we combined genome-wide siRNA analyses with interrogation of human interactome databases to assemble a host-pathogen biochemical network containing 213 confirmed host cellular factors and 11 HIV-1-encoded proteins. Protein complexes that regulate ubiquitin conjugation, proteolysis, DNA-damage response, and RNA splicing were identified as important modulators of early-stage HIV-1 infection. Additionally, over 40 new factors were shown to specifically influence the initiation and/or kinetics of HIV-1 DNA synthesis, including cytoskeletal regulatory proteins, modulators of posttranslational modification, and nucleic acid-binding proteins. Finally, 15 proteins with diverse functional roles, including nuclear transport, prostaglandin synthesis, ubiquitination, and transcription, were found to influence nuclear import or viral DNA integration. Taken together, the multiscale approach described here has uncovered multiprotein virus-host interactions that likely act in concert to facilitate the early steps of HIV-1 infection.
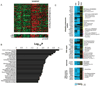
Figures




References
-
- Arhel N, Genovesio A, Kim KA, Miko S, Perret E, Olivo-Marin JC, Shorte S, Charneau P. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat Methods. 2006;3:817–824. - PubMed
-
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of Host Proteins Required for HIV Infection Through a Functional Genomic Screen. Science. 2008 - PubMed
-
- Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

