Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization
- PMID: 18854163
- PMCID: PMC2989779
- DOI: 10.1016/j.cell.2008.07.049
Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization
Abstract
Drosophila neural precursor cells divide asymmetrically by segregating the Numb protein into one of the two daughter cells. Numb is uniformly cortical in interphase but assumes a polarized localization in mitosis. Here, we show that a phosphorylation cascade triggered by the activation of Aurora-A is responsible for the asymmetric localization of Numb in mitosis. Aurora-A phosphorylates Par-6, a regulatory subunit of atypical protein kinase C (aPKC). This activates aPKC, which initially phosphorylates Lethal (2) giant larvae (Lgl), a cytoskeletal protein that binds and inhibits aPKC during interphase. Phosphorylated Lgl is released from aPKC and thereby allows the PDZ domain protein Bazooka to enter the complex. This changes substrate specificity and allows aPKC to phosphorylate Numb and release the protein from one side of the cell cortex. Our data reveal a molecular mechanism for the asymmetric localization of Numb and show how cell polarity can be coupled to cell-cycle progression.
Figures
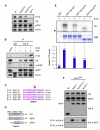
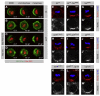
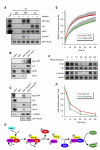
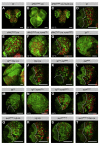
 denotes any hydrophobic residue. (D) Constructs used in the kinase assay. (E) AurA phosphorylates Par-6 on Ser34 in vitro. Recombinant proteins were incubated with [32P]ATP and recombinant AurA as indicated. The gel was Coomassie-stained (CBB), followed by autoradiography (32P). The arrowhead indicates autophosphorylated AurA. Autoradiographs were quantified by summing the signal of the full-length bands and normalizing for the signal of Par-6WT (set to 1). Averages and standard deviations are shown (n = 4). (F) AurA phosphorylates Par-6 on Ser34 in vivo. Phosphospecific antibodies directed against Ser34 were used to immunoprecipitate p-Par-6 from brains of the indicated genotypes. A second round (#2) of immunoprecipitation from the supernatant confirms the depletion of p-Par-6. To avoid the overlapping IgG signal and to control for the specificity of the antibodies, extracts from wild-type animals and from par-6Δ226 mutants complemented by a genomic rescue construct expressing Par-6-GFP (par-6GFP) were used.
denotes any hydrophobic residue. (D) Constructs used in the kinase assay. (E) AurA phosphorylates Par-6 on Ser34 in vitro. Recombinant proteins were incubated with [32P]ATP and recombinant AurA as indicated. The gel was Coomassie-stained (CBB), followed by autoradiography (32P). The arrowhead indicates autophosphorylated AurA. Autoradiographs were quantified by summing the signal of the full-length bands and normalizing for the signal of Par-6WT (set to 1). Averages and standard deviations are shown (n = 4). (F) AurA phosphorylates Par-6 on Ser34 in vivo. Phosphospecific antibodies directed against Ser34 were used to immunoprecipitate p-Par-6 from brains of the indicated genotypes. A second round (#2) of immunoprecipitation from the supernatant confirms the depletion of p-Par-6. To avoid the overlapping IgG signal and to control for the specificity of the antibodies, extracts from wild-type animals and from par-6Δ226 mutants complemented by a genomic rescue construct expressing Par-6-GFP (par-6GFP) were used.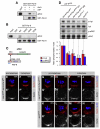
References
-
- Barros CS, Phelps CB, Brand AH. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev. Cell. 2003;5:829–840. - PubMed
-
- Bellaiche Y, Gho M, Kaltschmidt JA, Brand AH, Schweisguth F. Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat. Cell Biol. 2001a;3:50–57. - PubMed
-
- Bellaiche Y, Radovic A, Woods DF, Hough CD, Parmentier ML, O’Kane CJ, Bryant PJ, Schweisguth F. The Partner of Inscuteable/Discs-large complex is required to establish planar polarity during asymmetric cell division in Drosophila. Cell. 2001b;106:355–366. - PubMed
-
- Benton R, St Johnston D. Drosophila PAR-1 and 14–3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. - PubMed
-
- Berdnik D, Knoblich JA. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr. Biol. 2002;12:640–647. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

