Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila
- PMID: 18854242
- PMCID: PMC2584229
- DOI: 10.1016/j.chom.2008.09.001
Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila
Abstract
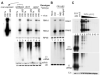
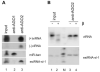
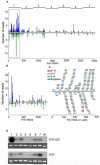
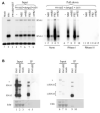
The small RNA-directed viral immunity pathway in plants and invertebrates begins with the production by Dicer nuclease of virus-derived siRNAs (viRNAs), which guide specific antiviral silencing by Argonaute protein in an RNA-induced silencing complex (RISC). Molecular identity of the viral RNA precursor of viRNAs remains a matter of debate. Using Flock house virus (FHV) infection of Drosophila as a model, we show that replication of FHV positive-strand RNA genome produces an approximately 400 bp dsRNA from its 5' terminus that serves as the major Dicer-2 substrate. ViRNAs thus generated are loaded in Argonaute-2 and methylated at their 3' ends. Notably, FHV-encoded RNAi suppressor B2 protein interacts with both viral dsRNA and RNA replicase and inhibits production of the 5'-terminal viRNAs. Our findings, therefore, provide a model in which small RNA-directed viral immunity is induced during the initiation of viral progeny (+)RNA synthesis and suppressed by B2 inside the viral RNA replication complex.
Figures

References
-
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. - PubMed
-
- Chapman EL, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Gene. 2007;7:844–96. - PubMed
-
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952–957. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

