Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation
- PMID: 18854817
- PMCID: PMC2583085
- DOI: 10.1038/msb.2008.59
Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation
Abstract
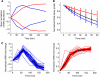
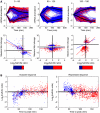
The state of the transcriptome reflects a balance between mRNA production and degradation. Yet how these two regulatory arms interact in shaping the kinetics of the transcriptome in response to environmental changes is not known. We subjected yeast to two stresses, one that induces a fast and transient response, and another that triggers a slow enduring response. We then used microarrays following transcriptional arrest to measure genome-wide decay profiles under each condition. We found condition-specific changes in mRNA decay rates and coordination between mRNA production and degradation. In the transient response, most induced genes were surprisingly destabilized, whereas repressed genes were somewhat stabilized, exhibiting counteraction between production and degradation. This strategy can reconcile high steady-state level with short response time among induced genes. In contrast, the stress that induces the slow response displays the more expected behavior, whereby most induced genes are stabilized, and repressed genes are destabilized. Our results show genome-wide interplay between mRNA production and degradation, and that alternative modes of such interplay determine the kinetics of the transcriptome in response to stress.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Garcia-Martinez J, Aranda A, Perez-Ortin JE (2004) Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell 15: 303–313 - PubMed
-
- Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

