Monitoring helicase activity with molecular beacons
- PMID: 18855770
- PMCID: PMC2575809
- DOI: 10.2144/000112834
Monitoring helicase activity with molecular beacons
Abstract
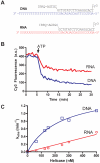
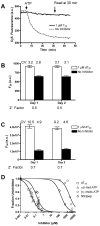
A high-throughput, fluorescence-based helicase assay using molecular beacons is described. The assay is tested using the NS3 helicase encoded by the hepatitis C virus (HCV) and is shown to accurately monitor helicase action on both DNA and RNA. In the assay, a ssDNA oligonucleotide molecular beacon, featuring a fluorescent moiety attached to one end and a quencher attached to the other, is annealed to a second longer DNA or RNA oligonucleotide. Upon strand separation by a helicase and ATP, the beacon strand forms an intramolecular hairpin that brings the tethered fluorescent and quencher molecules into juxtaposition, quenching fluorescence. Unlike currently available real-time helicase assays, the molecular beacon-based helicase assay is irreversible. As such, it does not require the addition of extra DNA strands to prevent products from re-annealing. Several variants of the new assay are described and experimentally verified using both Cy3 and Cy5 beacons, including one based on a sequence from the HCV genome. The HCV genome-based molecular beacon helicase assay is used to demonstrate how such an assay can be used in high-throughput screens and to analyze HCV helicase inhibitors.
Figures



Similar articles
-
Identification and analysis of inhibitors targeting the hepatitis C virus NS3 helicase.Methods Enzymol. 2012;511:463-83. doi: 10.1016/B978-0-12-396546-2.00021-8. Methods Enzymol. 2012. PMID: 22713333 Free PMC article.
-
Fuel specificity of the hepatitis C virus NS3 helicase.J Mol Biol. 2009 May 15;388(4):851-64. doi: 10.1016/j.jmb.2009.03.059. Epub 2009 Mar 28. J Mol Biol. 2009. PMID: 19332076 Free PMC article.
-
Real-time monitoring of RNA helicase activity using fluorescence resonance energy transfer in vitro.Biochem Biophys Res Commun. 2010 Feb 26;393(1):131-6. doi: 10.1016/j.bbrc.2010.01.100. Epub 2010 Feb 1. Biochem Biophys Res Commun. 2010. PMID: 20117090
-
Structure and function of hepatitis C virus NS3 helicase.Curr Top Microbiol Immunol. 2000;242:171-96. doi: 10.1007/978-3-642-59605-6_9. Curr Top Microbiol Immunol. 2000. PMID: 10592661 Review.
-
The hepatitis C virus NS3 protein: a model RNA helicase and potential drug target.Curr Issues Mol Biol. 2007 Jan;9(1):1-20. Curr Issues Mol Biol. 2007. PMID: 17263143 Free PMC article. Review.
Cited by
-
Label-free luminescence switch-on detection of hepatitis C virus NS3 helicase activity using a G-quadruplex-selective probe.Chem Sci. 2015 Apr 1;6(4):2166-2171. doi: 10.1039/c4sc03319a. Epub 2014 Nov 25. Chem Sci. 2015. PMID: 28808523 Free PMC article.
-
Structural basis for MTR4-ZCCHC8 interactions that stimulate the MTR4 helicase in the nuclear exosome-targeting complex.Proc Natl Acad Sci U S A. 2018 Jun 12;115(24):E5506-E5515. doi: 10.1073/pnas.1803530115. Epub 2018 May 29. Proc Natl Acad Sci U S A. 2018. PMID: 29844170 Free PMC article.
-
Identification and analysis of hepatitis C virus NS3 helicase inhibitors using nucleic acid binding assays.Nucleic Acids Res. 2012 Sep 1;40(17):8607-21. doi: 10.1093/nar/gks623. Epub 2012 Jun 27. Nucleic Acids Res. 2012. PMID: 22740655 Free PMC article.
-
Helicase inhibitors as specifically targeted antiviral therapy for hepatitis C.Future Virol. 2009 May 1;4(3):277-293. doi: 10.2217/fvl.09.7. Future Virol. 2009. PMID: 20161209 Free PMC article.
-
A new helicase assay based on graphene oxide for anti-viral drug development.Mol Cells. 2013 Apr;35(4):269-73. doi: 10.1007/s10059-013-0066-1. Epub 2013 Mar 11. Mol Cells. 2013. PMID: 23483279 Free PMC article. Review.
References
-
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. - PubMed
-
- Matson SW, Tabor S, Richardson CC. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J. Biol. Chem. 1983;258:14017–14024. - PubMed
-
- Sivaraja M, Giordano H, Peterson MG. High-throughput screening assay for helicase enzymes. Anal. Biochem. 1998;265:22–27. - PubMed
-
- Earnshaw DL, Pope AJ. FlashPlate scintillation proximity assays for characterization and screening of DNA polymerase, primase, and helicase activities. J Biomol Screen. 2001;6:39–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
