Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner
- PMID: 18922021
- PMCID: PMC2650222
- DOI: 10.1021/bi8015982
Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner
Abstract
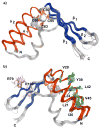
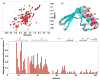
RNase P is the ubiquitous ribonucleoprotein metalloenzyme responsible for cleaving the 5'-leader sequence of precursor tRNAs during their maturation. While the RNA subunit is catalytically active on its own at high monovalent and divalent ion concentrations, four protein subunits are associated with archaeal RNase P activity in vivo: RPP21, RPP29, RPP30, and POP5. These proteins have been shown to function in pairs: RPP21-RPP29 and POP5-RPP30. We have determined the solution structure of RPP21 from the hyperthermophilic archaeon Pyrococcus furiosus ( Pfu) using conventional and paramagnetic NMR techniques. Pfu RPP21 in solution consists of an unstructured N-terminus, two alpha-helices, a zinc binding motif, and an unstructured C-terminus. Moreover, we have used chemical shift perturbations to characterize the interaction of RPP21 with RPP29. The data show that the primary contact with RPP29 is localized to the two helices of RPP21. This information represents a fundamental step toward understanding structure-function relationships of the archaeal RNase P holoenzyme.
Figures
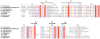
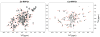

References
-
- Altman S, Baer MF, Bartkiewicz M, Gold H, Guerrier-Takada C, Kirsebom LA, Lumelsky N, Peck K. Catalysis by the RNA subunit of RNase P--a minireview. Gene. 1989;82:63–64. - PubMed
-
- Gopalan V, Altman S. In: Ribonuclease P: Structure and Catalysis, in The RNA World. Gesteland R, Cech T, Atkins J, editors. Cold Spring Harbor Laboratory Press; New York: 2006. only online at http://rna.cshl.edu)
-
- Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem Sci. 2006;31:333–341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

