An investigation of the statistical power of neutrality tests based on comparative and population genetic data
- PMID: 18922762
- PMCID: PMC2727393
- DOI: 10.1093/molbev/msn231
An investigation of the statistical power of neutrality tests based on comparative and population genetic data
Abstract
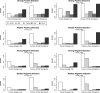
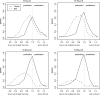
In this report, we investigate the statistical power of several tests of selective neutrality based on patterns of genetic diversity within and between species. The goal is to compare tests based solely on population genetic data with tests using comparative data or a combination of comparative and population genetic data. We show that in the presence of repeated selective sweeps on relatively neutral background, tests based on the d(N)/d(S) ratios in comparative data almost always have more power to detect selection than tests based on population genetic data, even if the overall level of divergence is low. Tests based solely on the distribution of allele frequencies or the site frequency spectrum, such as the Ewens-Watterson test or Tajima's D, have less power in detecting both positive and negative selection because of the transient nature of positive selection and the weak signal left by negative selection. The Hudson-Kreitman-Aguadé test is the most powerful test for detecting positive selection among the population genetic tests investigated, whereas McDonald-Kreitman test typically has more power to detect negative selection. We discuss our findings in the light of the discordant results obtained in several recently published genomic scans.
Figures
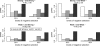

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

