Interaction of the RAGE cytoplasmic domain with diaphanous-1 is required for ligand-stimulated cellular migration through activation of Rac1 and Cdc42
- PMID: 18922799
- PMCID: PMC2590709
- DOI: 10.1074/jbc.M801465200
Interaction of the RAGE cytoplasmic domain with diaphanous-1 is required for ligand-stimulated cellular migration through activation of Rac1 and Cdc42
Abstract
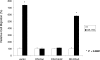
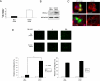
Cellular migration is a fundamental process linked to diverse pathological states such as diabetes and its complications, atherosclerosis, inflammation, and cancer. The receptor for advanced glycation end products (RAGE) is a multiligand cell surface macromolecule which binds distinct ligands that accumulate in these settings. RAGE-ligand interaction evokes central changes in key biological properties of cells, including proliferation, generation of inflammatory mediators, and migration. Although RAGE-dependent signal transduction is critically dependent on its short cytoplasmic domain, to date the proximate mechanism by which this RAGE domain engages and stimulates cytoplasmic signaling pathways has yet to be identified. Here we show that the RAGE cytoplasmic domain interacts with Diaphanous-1 (Dia-1) both in vitro and in vivo. We employed the human RAGE cytoplasmic domain as "bait" in the yeast two-hybrid assay and identified the formin homology (FH1) domain of Dia-1 as a potential binding partner of this RAGE domain. Immunoprecipitation studies revealed that the RAGE cytoplasmic domain interacts with the FH1 domain of Dia-1. Down-regulation of Dia-1 expression by RNA interference blocks RAGE-mediated activation of Rac-1 and Cdc42 and, in parallel, RAGE ligand-stimulated cellular migration. Taken together, these findings indicate that the interaction of the RAGE cytoplasmic domain with Dia-1 is required to transduce extracellular environmental cues evoked by binding of RAGE ligands to their cell surface receptor, a chief consequence of which is Rac-1 and Cdc42 activation and cellular migration. Because RAGE and Dia-1 are implicated in the regulation of inflammatory, vascular, and transformed cell migration, these findings highlight this interaction as a novel target for therapeutic intervention in inflammation, atherosclerosis, diabetes, and cancer.
Figures







References
-
- Neeper, M., Schmidt, A. M., Brett, J., Yan, S. D., Wang, F., Pan, Y. C. P., Elliston, K., Stern, D., and Shaw, A. (1992) J. Biol. Chem. 267 14998-15004 - PubMed
-
- Hofmann, M. A., Drury, S., Fu, C. F., Qu, W., Taguchi, A., Lu, Y., Avila, C., Kambham, N., Bierhaus, A., Nawroth, P., Neurath, M. F., Slattery, T., Beach, D., McClary, J., Nagashima, M., Morser, J., Stern, D., and Schmidt, A. M. (1999) Cell 97 889-901 - PubMed
-
- Hori, O., Brett, J., Slattery, T., Cao, R., Zhang, J., Chen, J. X., Nagashima, M., Lundh, E. R., Vijay, S., Nitecki, D., Morser, J., Stern, D., and Schmidt, A. M. (1995) J. Biol. Chem. 270 25752-25761 - PubMed
-
- Yan, S. D., Chen, X., Fu, J., Chen, M., Zhu, H., Roher, A., Slattery, T., Zhao, L., Nagashima, M., Morser, J., Migheli, A., Nawroth, P., Stern, D., and Schmidt, A. M. (1996) Nature 382 685-691 - PubMed
-
- Huttunen, H. J., Fages, C., and Rauvala, H. (1999) J. Biol. Chem. 274 19919-19924 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

